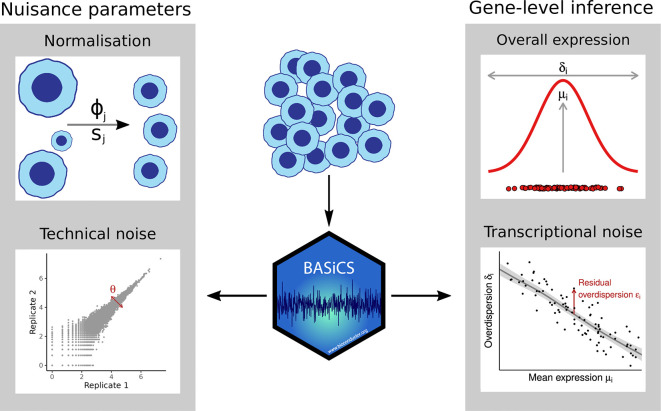
Figure 1. A schematic representation of the features of the BASiCS model.
BASiCS accounts for cell-to-cell and batch-to-batch variability using nuisance parameters. Accounting for cell-to-cell and batch-to-batch technical variability allows BASiCS to robustly perform gene-level inference of mean and variability. Furthermore, by accounting for the association between mean and variability in scRNAseq, BASiCS can also infer transcriptional noise using the residual variability parameter ε i .