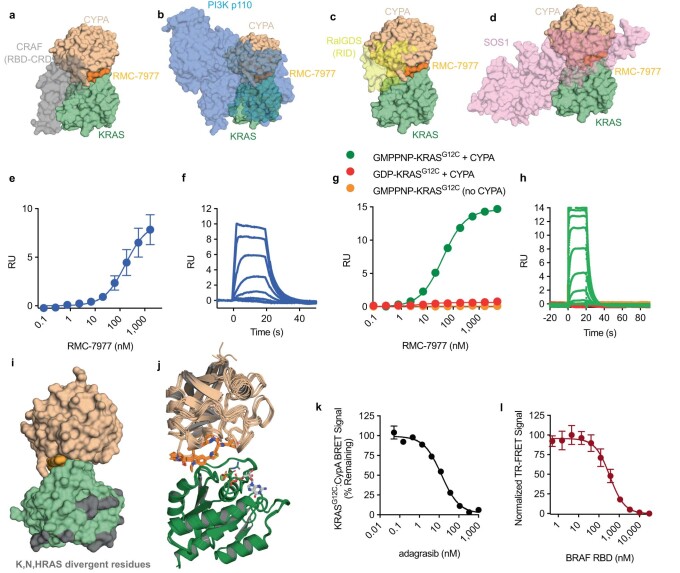
Extended Data Fig. 2. RMC-7977 biophysical and structural characterization.
a,b,c,d, Superimposition of the CYPA:RMC-7977:KRAS wild-type (WT) tri-complex structure (PDB: 8TBF) with KRAS:CRAF RBD-CRD complex (a, PDB: 6XI7), HRAS:PIK3 CD (b, PDB: 1HE8), HRAS:RALGDS (c, PDB: 1LFD), or KRAS:SOS1 REM-CDC25 (d, PDB: 1XD2). Note steric clashes caused by CYPA occupying the Switch I and II motif of KRAS. e,f, Steady state (e) and kinetic sensorgram (f) of RMC-7977 and CYPA binding measured by SPR response units (RU). g,h, Steady state (g) and kinetic sensorgram (h) of RMC-7977 binding to KRASG12C measured by SPR. Measurements taken in the presence or absence of CYPA, and GMPPNP or GDP nucleotides. e,g, Datapoints represent mean ±s.d. of 3 biological replicates. i, RAS interacts with CYPA and RMC-7977 through the conserved effector lobe. Isoform divergent residues are distal from the site of interaction, allowing pan-isoform inhibition. j, Alignment of wild-type HRAS, wild-type NRAS, and KRASG12X (A, C, D, R, S, V) mutant tri-complexes shows functionally identical binding modes to the CYPA:RMC-7977 binary complex. 12th position mutant sidechains shown as sticks (PDB: 8TBF, 8TBG, 8TBH, 8TBI, 8TBJ, 8TBK, 8TBL, 8TBM, 8TBN). k, Tri-complex formation assay (KRASG12C:RMC-7977:CYPA binding in U2OS cells) in the presence of 100 nM RMC-7977 and the indicated concentration of adagrasib, normalized to the expected min. (100 nM RMC-7977, no adagrasib) and max. (0 nM RMC-7977, no adagrasib) BRET signal. Cells were treated with inhibitor for 4 h. Datapoints represent mean ±s.d. of 6 biological replicates. l, Time resolved-FRET between recombinant KRASG12V and CYPA in the presence of RMC-7977 (50 nM of each) treated with the indicated concentrations of recombinant BRAF RBD, normalized as % of DMSO (EC50 = 357 nM, 95% CI = 270-505 nM). n = 3 biological replicates, plotted as mean ±s.d. normalized to control.