Figure 4.
FACS isolation and qPCR of MolTimer2.0 expressing neural progenitor cells
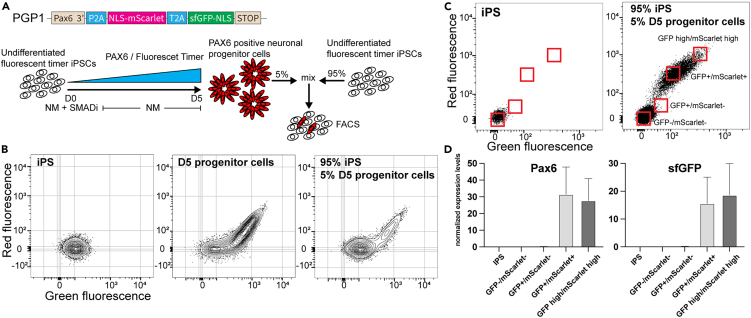
(A) Schematic of the mixing and FACS isolation of MolTimer2.0 expressing neural progenitor cells. PGP1-PAX6-MolTimer2.0 neural progenitors were mixed with undifferentiated PGP1-PAX6-MolTimer2.0 iPSC prior to FACS analysis.
(B) FACS plots of PGP1-PAX6-MolTimer2.0 iPSC (left), D5 PGP1-PAX6-MolTimer2.0 neural progenitor cells (middle), and the 1:20 mix of D5 PGP1-PAX6-MolTimer2.0 progenitor cells and undifferentiated PGP1-PAX6-MolTimer2.0 iPSCs used for isolation and qPCR analysis (right).
(C) Gating strategy to isolate four distinct populations of MolTimer2.0 expressing neural progenitor cells: non-fluorescent GFP−/mScarlet−, green-only GFP+/mScarlet−, dual fluorescent GFP+/mScarlet+, and high level dual fluorescent GFP+/mScarlet+.
(D) qPCR analysis of PAX6 and sfGFP expression. PAX6 and sfGFP are only detected in the low level GFP+/mScarlet+ and high level GFP+/mScarlet+ populations. Data are represented as mean ± SD.