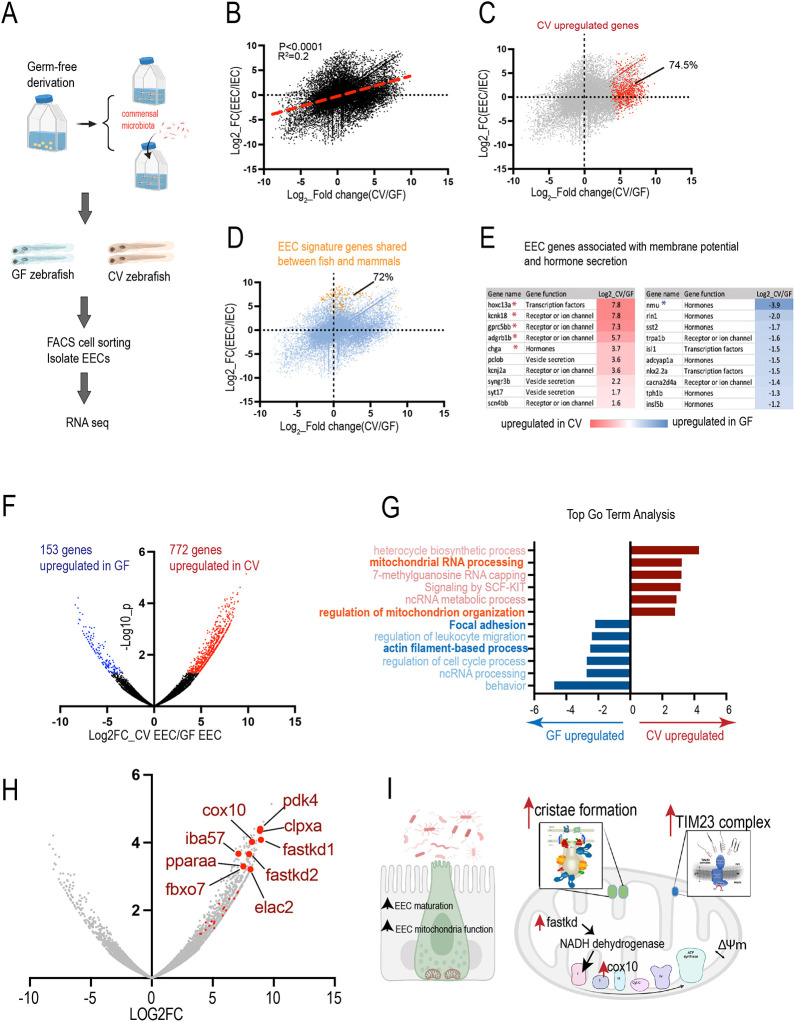
Fig. 3.
Gut microbiota promote EEC maturation and mitochondrial function. (A) Experimental overview for transcriptomic analysis of the FACS-sorted EECs from 8 dpf Tg(neurod1:RFP); Tg(cldn15la:EGFP) GF and CV zebrafish. Tg(cldn15la:EGFP) labels the intestine epithelium and Tg(neurod1:RFP) labels the EECs in the intestinal epithelium. Cells with both GFP and RFP fluorescence were sorted. (B) Positive correlation between the genes that are upregulated in CV (x-axis) and the genes that are enriched in EECs (y-axis). (C) Among the genes that are significantly upregulated in CV (red), 74.5% are enriched in the EECs. (D) Of the EECs signature genes shared between zebrafish and mammals, 72% are upregulated in CV. (E) Differential expression of the EEC signature genes that encode hormone peptides or are involved in membrane potential in GF and CV conditions. Asterisks indicate that the genes are significantly upregulated in the GF or CV conditions. (F) Volcano plot showing the genes that are significantly upregulated in CV or GF. (G) GO term analysis of the CV or GF upregulated genes. (H) Volcano plot showing the genes that are involved in mitochondrial function. Many of the genes that are associated with mitochondrial regulation are among the most significantly upregulated genes in CV EECs. (I) Model figure showing that commensal microbiota colonization promotes EEC maturation and mitochondrial function.