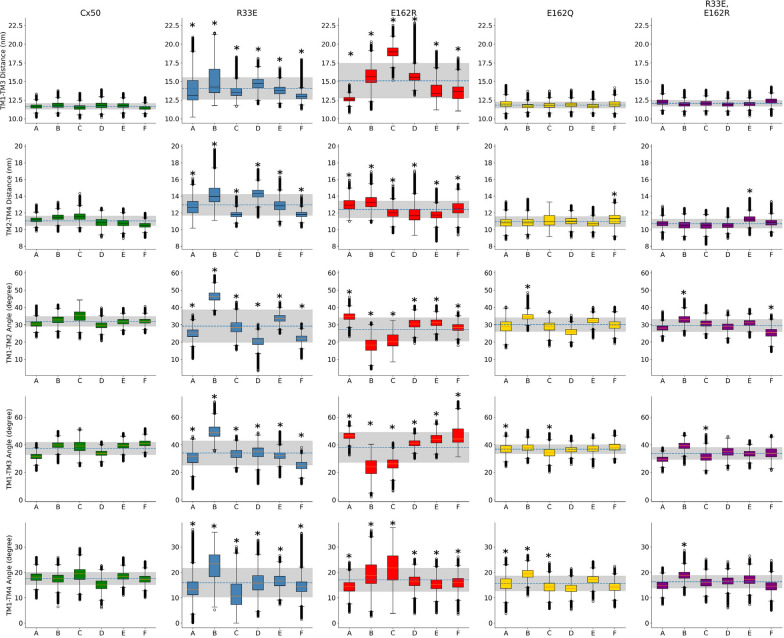
Fig. 4.
Distance and angle variations between the transmembrane domains of the mutant protomers compared with the wild-type Cx50 protomer. Graphs show the distance (top two rows) and the angle (bottom 3 rows) between transmembrane domains in Cx50, Cx50R33E, Cx50E162R, Cx50E162Q and Cx50R33E,E162R protomers. The distance between TM1 and TM3 was measured between amino acid residues 25 and 159, and that between TM2 and TM4 was measured between amino acid residues 85 and 220. The inclination of TM2, TM3 and TM4 with respect to TM1 was calculated by finding the best fitted straight line passing through the Cα of all amino acid residues in each transmembrane domain. The results are presented as box plots (mean ± S.D.) for each protomer (A‒F) using the measurement from all frames from all three replicas. The outliers are presented as empty circles. The horizontal dashed line corresponds to the average for all six protomers for each construct and the gray shade corresponds to the standard deviation. Asterisks indicate a significant difference vs. the corresponding monomer in wild-type Cx50 using Kruskal–Wallis test (p < 0.05)