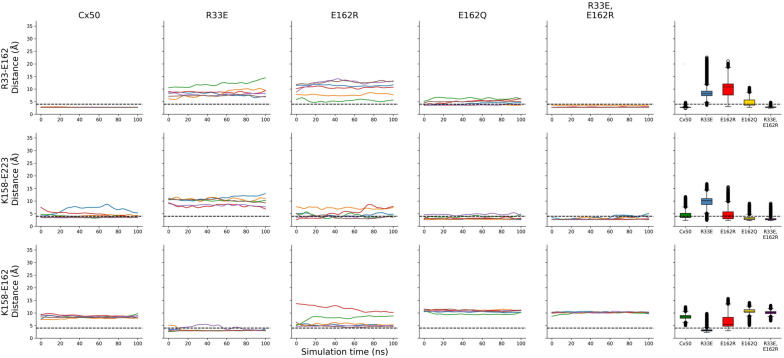
Fig. 5.
Amino acids within the intracellular pocket involved in salt-bridge formation. Graphs show the distance between selected pairs of amino acid residues: positions 33–162, positions 158-223, and positions 158-162 in wild-type and mutant Cx50. Distances were calculated from the center of mass of the terminal moieties of each residue (guanidinium for R; carboxylate for E; amide for Q). The graphs in the first five columns show the time-course of variation of the mean value of the distance in the three 100-ns simulation replicas for each protomer (plotted in a different color). The last column shows box plots of the same distances, with all protomers analyzed together. The black dashed line marks the 4 Å maximum distance limit above which two charged residues could not form a salt bridge