Fig. 2.
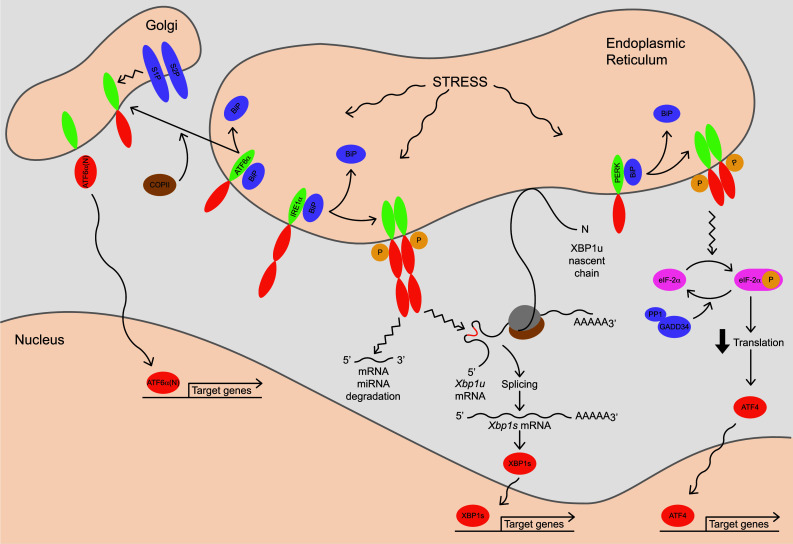
Signaling pathways of the mammalian UPR. Conditions that increase demand on the ER induce ‘stress’ and trigger the UPR. Upon release from the chaperone BiP, an exposed Golgi-localization signal and interaction with the COPII complex allows ATF6α to be ferried to the Golgi apparatus where it undergoes regulated intramembrane proteolysis by S1P and S2P. Liberated from the membrane, its cytosolic domain, ATF6α(N), moves into the nucleus where it functions as a bZIP transcription factor to up-regulate expression of genes via the ERSE promoter motif. ATF6α primarily targets genes involved in various ER quality control processes such as protein folding and ERAD. The IRE1α proteins oligomerize when BiP releases, activating their C-terminal endoribonuclease domains that execute site-specific cleavage of Xbp1 mRNA at two sites. Splicing of the resulting fragments in the cytoplasm yields a transcript with an altered reading frame encoding XBP1s, a bZIP transcription factor that acts on the ERSE and UPRE promoter motifs to up-regulate genes involved throughout the secretory pathway. Translation of Xbp1 mRNA prior to UPR-directed splicing yields XBP1u, a bZIP factor which lacks a transactivation domain. A hydrophobic region near the C-terminus of nascent XBP1u associates with the ER membrane. This interaction facilitates IRE1α-initiated splicing as it positions ribosome-engaged Xbp1 mRNA near the ER membrane. Activated IRE1α also cleaves and degrades select mRNA and miRNA. Upon release of BiP, the PERK proteins oligomerize and phosphorylate the translation initiation factor eIF-2α, effectively reducing translation. Translation of ATF4 increases when global protein synthesis decreases. ATF4 acts on the CARE promoter motif to induce a variety of targets including genes involved in cellular redox homeostasis, amino acid metabolism, protein synthesis and apoptosis. The GADD34–PP1 complex de-phosphorylates eIF-2α, allowing translation to resume
