Fig. 2.
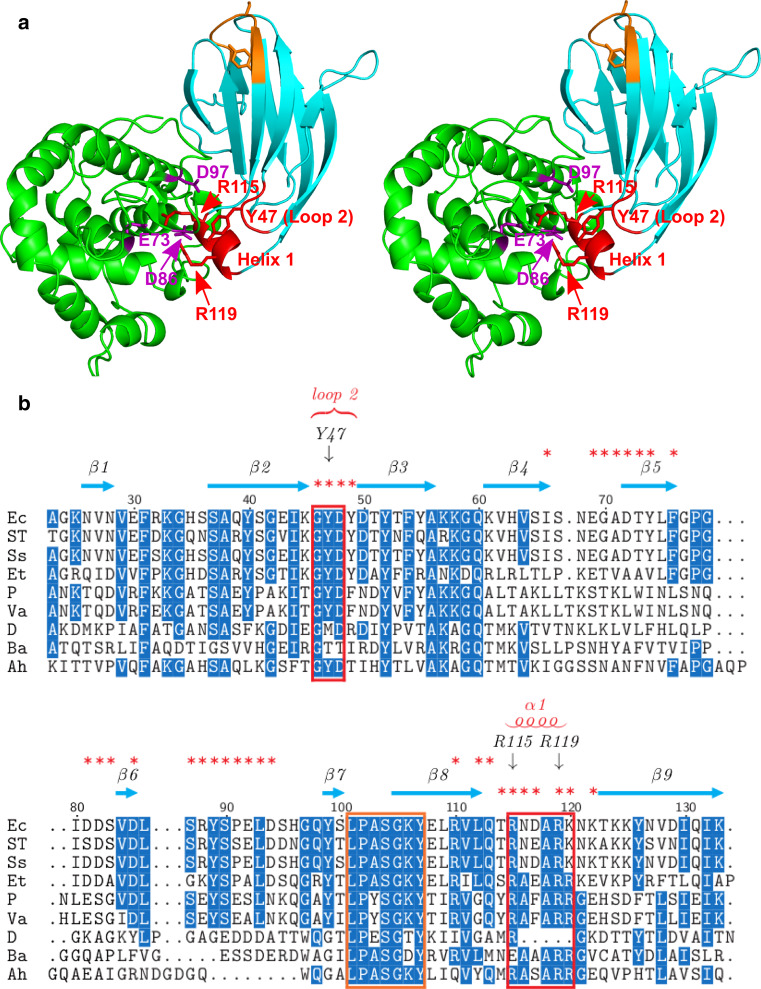
Two conserved regions of PliG-Ec interact with the SalG active site. a Stereo view of SalG (green) and PliG-Ec (cyan) in the complex. PliG-Ec residues Y47 in loop 2 of as well as residues R115 and R119 in helix 1 (red labels) interact with E73, D86 and D97 (purple labels) in the active site of SalG. These residues are located in two highly conserved regions (indicated by red boxes in b). Another conserved region of PliG-Ec (marked with orange here and in b) contains the SGxY sequence motif shown to be important for lysozyme inhibition in the PliC/MliC and PliI families. The SalG loop containing residue D86 is shown here in the ‘inward’ conformation (see Fig. 4 for more detail). b Multiple sequence alignment of PliG homologues. Ec, E. coli (NP_287417.1), ST S. Typhimurium (ACY87940.1), Ss Shigella sonnei (YP_310126.1), Et Edwardsiella tarda (YP_003296035.1), P Photobacterium sp. SKA34 (ZP_01159524.1), Va Vibrio angustum (ZP_01234798.1), D Desulfovibrio sp. FW1012B (ZP_06368489.1), Ba B. avium (YP_785764.1), Ah A. hydrophila (YP_854659.1). Red asterisks mark all PliG-Ec residues that are located on the SalG/PliG-Ec interface. The alignment was created with STRAP [48]