Abstract
MicroRNAs (miRNAs), a novel class of molecules regulating gene expression, have been hailed as modulators of many biological processes and disease states. Recent studies demonstrated an important role of miRNAs in the processes of inflammation and cancer, however, there are little data implicating miRNAs in peripheral pain. Bladder pain syndrome/interstitial cystitis (BPS/IC) is a clinical syndrome of pelvic pain and urinary urgency/frequency in the absence of a specific cause. BPS is a chronic inflammatory condition that might share some of the pathogenetic mechanisms with its common co-morbidities inflammatory bowel disease (IBD), asthma and autoimmune diseases. Using miRNA profiling in BPS and the information about validated miRNA targets, we delineated the signaling pathways activated in this and other inflammatory pain disorders. This review projects the miRNA profiling and functional data originating from the research in bladder cancer and immune-mediated diseases on the BPS-specific miRNAs with the aim to gain new insight into the pathogenesis of this enigmatic disorder, and highlighting the common regulatory mechanisms of pain and inflammation.
Keywords: MicroRNA, Bladder, Pain, Inflammation, Gene expression, Bladder cancer, Inflammatory bowel disease
Introduction
MicroRNAs (miRNAs) are quickly gaining recognition for their role in many biological processes and disease states [1, 2]. MiRNAs are endogenous non-coding single-stranded RNAs of approximately 22 nucleotides that regulate gene expression by post-transcriptional mechanisms upon sequence-specific binding to the 3′ untranslated regions (3′ UTRs) [1, 3] or, occasionally, the 5′UTRs [4, 5] or coding regions [6–8] of their mRNA targets. MiRNAs exhibit imperfect complementarity with mRNAs, allowing them to regulate multiple genes and thus complicating efforts to predict and functionally validate their targets [9]. Since their discovery [10], miRNAs have been acknowledged as important modulators of gene expression, and deficiency in their synthesis or function contributes to many human diseases [11, 12]. MiRNA expression profiles characteristic of a particular disorder may serve as a useful diagnostic tool, but more importantly, it is the first step towards unraveling miRNA functions. Follow-up studies in experimental models are carried out to identify the protein targets of differentially expressed miRNAs and delineate the signaling pathways activated in a diseased state.
Although a wealth of information has been gathered on miRNA expression in bladder cancer (BCa), which in this respect remains the best-studied disorder, only a few studies have been carried out in bladder dysfunction. The first miRNA profiling in bladder pain syndrome/interstitial cystitis (BPS), which was performed in our laboratory, has identified several miRNAs, regulating the expression of signaling and adhesion molecules [13, 14]. Pain and inflammation are characteristic of BPS, which has been suggested to share the pathogenetic mechanisms with other inflammatory diseases such as asthma, inflammatory bowel disease, and autoimmune diseases [15]. These are common co-morbidities of BPS/IC, and miRNA expression and function in these immune-mediated diseases has been addressed in a number of studies. This review projects the miRNA profiling and functional data originating from the research in bladder cancer and immune-mediated diseases on the BPS-specific miRNAs aiming to gain new insight into the pathogenesis of this enigmatic disorder, and highlighting the common regulatory mechanisms of pain and inflammation.
MicroRNAs—general information
The first miRNAs lin-4 and let-7 were discovered as important regulators of the normal temporal control of diverse postembryonic developmental events in C. elegans [10, 16]. Most miRNA reduce protein synthesis, whereas some, such as miR369-3 and miR-373, enhance translation [17, 18]. The first link between miRNAs and cancer was made after uncovering the deletion of miR-15 and miR-16 in leukemias [19], and the potential for the miRNA profiling in cancer diagnosis became apparent [20]. This was followed by studies of the role of miRNAs in cardiovascular [21], neurodegenerative [22–24], and autoimmune disease [25, 26]. Therapeutic use of miRNAs was pioneered by administration of miRNAs [27], or their antagonists called antagomirs [28], to the affected cells restoring the normal levels of their protein targets. The locked nucleic acid (LNA)-antagomirs have been successfully introduced into the clinical practice: LNA-anti-miR-122 suppresses hepatitis C virus (HCV) replication in chronically infected animals [29].
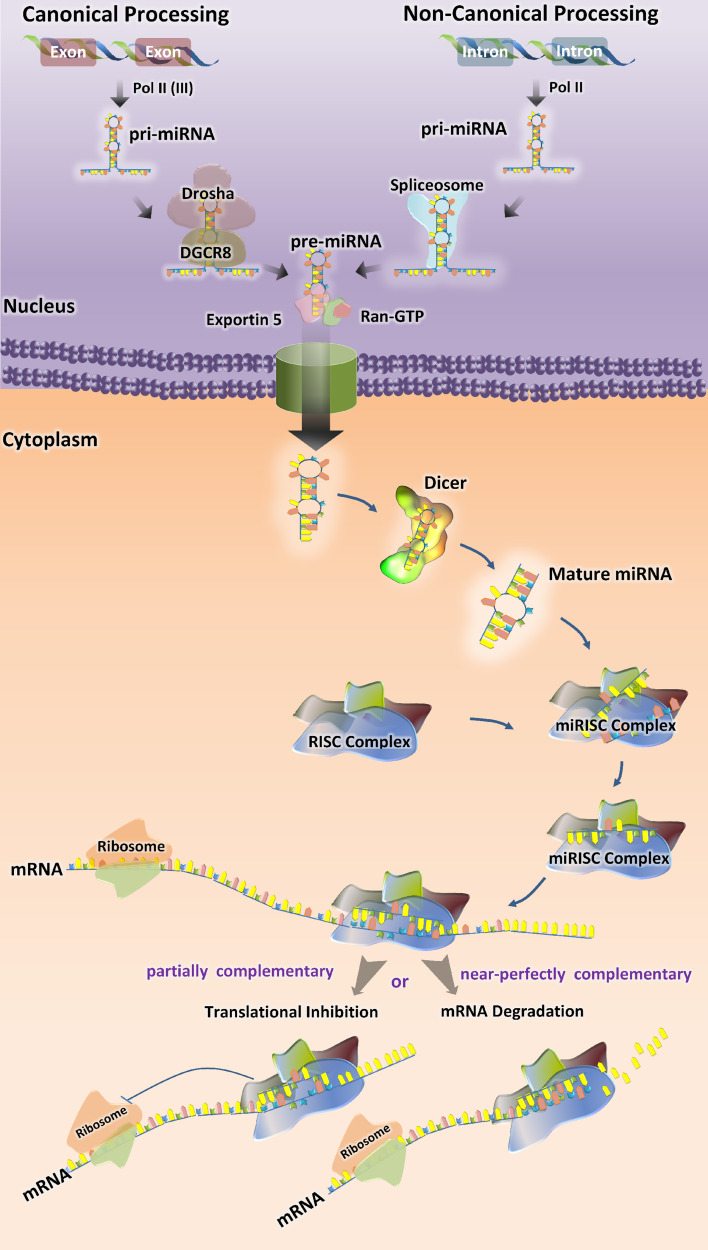
The details of miRNA synthesis are well characterized and extensively reviewed [30–32]. Most canonical mammalian miRNA genes have been identified in introns of the protein-coding or noncoding RNAs and around one-third of them are located in the introns of target genes [33]. The miRNA gene transcripts are the primary precursor RNAs (pri-miRNA), which mature in two steps (Fig. 1). Pri-miRNAs are processed in the nucleus by Drosha–DGCR8 complex into a ~70-nucleotide RNA molecules [34]. These precursor hairpin miRNAs (pre-miRNAs) are then exported to the cytoplasm by Exportin-5 (Exp5) [35] and cleaved by Dicer-TRBP complex into a ~20-bp miRNA/miRNA* duplex. The guide strand of the miRNA/miRNA* duplex is usually incorporated into miRNA-induced silencing complex (miRISC) and the passenger strand or (miRNA*) is released and degraded [36]. The final product is RISC-loaded mature miRNA, which is guided to its target mRNA by means of base pairing (Fig. 1).
Fig. 1.
Biogenesis of miRNAs. Mature microRNAs (miRNAs) are approximately 22 nucleotides long and generated by a two-step process. The first step takes place in the nucleus, where primary miRNAs (pri-miRNA) are converted into precursor miRNAs (pre-miRNAs) by the microprocessor complex containing Drosha and DGCR8 (in canonical process) or spliceosome, which excises the pri-microRNA transcribed from introns (in non-canonical process). Exportin-5 transports pre-miRNAs to the cytoplasm. In the second step, the hairpin structures of the pre-miRNAs are re-cleaved by the RNase III Dicer to generate mature miRNA. The functional strand of the mature miRNA gets incorporated into the RNA-induced Silencing Complex RISC (miRISC), where it guides miRISC to silence target mRNAs through translational repression or mRNA cleavage
Some microRNA, such as miR-877 and miR-1226, derive from short intronic hairpins named “mirtrons” [37, 38]. Mirtrons are spliced from the host gene by the spliceosome, become linearized by debranching enzyme and then fold into hairpins, which enter the miRNA-processing pathway without Drosha-mediated cleavage [37, 39]. Recently, a third pathway of microRNA biogenesis has been described: some microRNAs (miR-1225 and miR-1228) do not need DGCR8, Dicer, Exportin-5, or Argonaute 2 for their biogenesis, which nevertheless involves Drosha. This class of microRNAs has been named “simtrons” [40]. The miRNA registry in humans includes 18,226 entries for hairpin precursor miRNAs, expressing 21,643 mature miRNA products [41, 42].
Role of miRNAs in urinary bladder function and pathology
Pathogenesis of bladder pain syndrome (BPS)
Bladder pain syndrome (BPS) is a clinical syndrome of pelvic pain and urinary urgency-frequency in the absence of a specific cause. According to the European Society for the Study of Interstitial Cystitis (ESSIC) proposal and European Association of Urology (EAU) guidelines [43], BPS is diagnosed on the basis of symptoms of pain associated with the urinary bladder, accompanied by at least one other symptom, such as day-time and/or night-time urinary frequency after exclusion of confounding symptoms. Overall, this disease, which has a significant impact on social and psychological well-being, affects approximately 1 million patients in the USA alone [44], with at least 230 confirmed cases per 100,000 females [45]. The etiology of BPS is unknown, and its treatment largely empiric. A multitude of pathogenetic mechanisms have been postulated ranging from neuroinflammatory to autoimmune or possibly infectious or toxic agents, but an inflammatory component is commonly thought to be involved. Epithelial damage has often been invoked: the mucinous layer of the healthy bladder is often compromised in patients with BPS/IC, as well as in some animal models [46, 47].
The integrity of the urothelium is indispensable for the healthy bladder [48]. Mechanical or chemical damage, as well as a bacterial infection can lead to a compromised urothelium allowing urinary solutes to penetrate into the interstitium [49]. The loss of epithelial integrity is a predominant histopathologic finding in biopsies from BPS patients [50]. In human BPS patients, the molecular markers for bladder permeability and proteoglycan core proteins are often down-regulated [51, 52]. Recently, we have shown that the mRNA levels of the tight junction (TJ) proteins ZO-1, JAM-1, occludin, and tight claudins, normally present in water impermeable epithelia and abundant in normal urothelium, were significantly down-regulated in bladder biopsies from BPS patients, indicating a compromised tight junction structure and possibly increased permeability of the urothelium [13]. Therefore it seems possible that urothelial damage is a preceding feature of this disorder and might be a causative factor in the pathogenesis of BPS. A decrease of E-cadherin mRNA levels, which we observed at mRNA level in BPS (Monastyrskaya, unpublished), was confirmed by recent findings of lower E-cadherin and ZO-1 expression in IC/BPS, but not in overactive (OAB) bladders, and suggesting the urothelial barrier function was compromised in BPS but not affected in the OAB bladder [53].
BPS is characterized by several other gene expression changes: tachykinin receptors NK1R and NK2R were significantly down-regulated and bradykinin B1 receptor, cannabinoid receptor CB1, and muscarinic receptors M3–M5 were up-regulated in BPS patients’ biopsies [13]. In addition, expression of acid-sensing channels, important for nociceptive pain, was altered in BPS: we detected an up-regulation of ASIC2a and ASIC3 mRNA, whereas ASIC1a remained unchanged [54]. These changes, in conjunction with deficiencies of urothelial barrier, might account for increased nociception in BPS patients.
MiRNAs in BPS and OAB
New information is emerging on the miRNA-mediated regulation of epithelial permeability, bladder contractility, and neurogenic inflammation in bladder dysfunction. While studying the gene expression changes characteristic of BPS, we observed a perplexing down-regulation of tachykinin receptors in the biopsies of BPS patients which prompted us to study its mechanisms [13]. Using cell-based models, we showed that prolonged exposure of NK1R to Substance P (SP) caused a decrease of NK1R mRNA levels and a concomitant increase of regulatory miRNAs miR-449b and miR-500. In the biopsies of BPS patients, the same miRNAs were significantly increased, suggesting that BPS promoted an attenuation of NK1R synthesis via activation of specific miRNAs. We confirmed this hypothesis by identifying 31 differentially expressed miRNAs in BPS patients and demonstrated a direct correlation between miR-449b, miR-500, miR-328, and miR-320 and a down-regulation of NK1R mRNA and/or protein levels. The results of the first miRNA profiling in biopsies of BPS patients are shown in Table 1.
Table 1.
Comparative analysis of miRNA expression profiles
| Comparison of miRNA profiles | |||||
|---|---|---|---|---|---|
| miRNA | BPS | BCa | IBD (CD, UC) | Asthma, RA, MS, SLE | Pain |
| let-7 g | Up |
Up after opioid treatment [109] Altered in complex regional pain syndrome (CRPS) [113] |
|||
| miR-23a | Up | Up [60] | Up [150] | Up in SMC remodeling/asthma [88] | |
| miR-23b | Up | Up [60] | Up [151] | Up in SMC remodeling/asthma [88] | Up in neuropathic pain [110] |
| miR-25 | Up | Up in SMC remodeling/asthma [88] | |||
| miR-26a | Up | Up in SMC remodeling/asthma [126] | |||
| miR-27a | Up | ||||
| miR-30e-5p | Up | ||||
| miR-95 | Up | ||||
| miR-130b | Up | Down in CRPS [113] | |||
| miR-133b | Up |
Up [61], Down [66] |
Up [103] | Down after morphine—role in pain [152] | |
| miR-148a | Down | Up in SLE [79] | |||
| miR-182 | Down | ||||
| miR-186 | Up | ||||
| miR-192 | Up | Up [104] | Down in CRPS [113] | ||
| miR-199a(-5p) | Up | Down [66] | Up [153] | ||
| miR-320(a) | Up | Up [61] | Down [151] | ||
| miR-324-3p | Up | Up [103] | Down in CRPS [113] | ||
| miR-328 | Up | ||||
| miR-342(-3p) | Up | ||||
| miR-379 | Up | ||||
| miR-422a | Up | Up in MS [94] | Down in CRPS [113] | ||
| miR-422b(378) | Up | ||||
| miR-449b | Up | ||||
| miR-485-5p | Up | ||||
| miR-493 | Down | ||||
| miR-500(*) | Up | ||||
| miR-502(-5p) | Up | ||||
| miR-511 | Up | ||||
| miR-572 | Up | Up in MS [94] | |||
| miR-594 | Up | Up [151] | |||
| miR-597 | Up | ||||
Occurrence and regulation of the miRNAs, altered in BPS, BCa bladder cancer, IBD inflammatory bowel disease CD Crohn’s disease, UC ulcerative colitis, asthma, RA rheumatoid arthritis, SLE systemic lupus erythematosus, MS multiple sclerosis and pain
Recently, using a mouse model with an induced deletion of Dicer, two groups examined the role of miRNAs in the regulation of bladder contractility. An induced smooth-muscle-specific Dicer knock-out resulted in significantly reduced levels of miRNAs, including miR-145, miR-143, miR-22, miR-125b-5p, and miR-27a, from detrusor preparations without mucosa. Deletion of Dicer resulted in a disturbed micturition pattern in vivo and reduced depolarization-induced pressure development in an isolated detrusor due to decreased levels of L-type Ca2+ channels [55]. In a similar study, the loss of Dicer exacerbated cyclophosphamide-induced bladder overactivity in mice, possibly because the miRNAs capable of targeting P2X mRNAs were impaired, leading to enhanced P2X receptor expression [56]. The authors describe an up-regulation of miR-34a and miR-25 in the mouse model of OAB. Interestingly, miR-25 is also elevated in BPS [13] (Table 1), pointing to the similarities of function of this miRNA in both disorders.
Our follow-up study of the miRNA function in BPS concerned the role of these molecules in the regulation of urothelial permeability [14]. We identified microRNA miR-199a-5p, which was increased in BPS patients’ biopsies, as an important regulator of intercellular junctions. Upon overexpression in urothelial cells it impaired correct tight junction formation and caused a permeability increase. MiR-199a-5p directly targeted mRNAs encoding LIN7C, ARHGAP12, PALS1, RND1, and PVRL1 and attenuated their expression levels to a similar extent. Laser microdissection revealed that miR-199a-5p was predominantly expressed in the bladder smooth muscle, but also detected in the mature bladder urothelium and primary urothelial cultures. In the urothelium, its expression can be up-regulated following activation of cAMP signaling pathways. Our results point to a possible link between miR-199a-5p expression and the control of urothelial permeability in bladder pain syndrome. Up-regulation of miR-199a-5p and concomitant down-regulation of its multiple targets might be detrimental for the establishment of a tight urothelial barrier, leading to chronic pain.
miRNA profiling in bladder cancer (BCa)
Except the studies discussed above, most of the information on the role of miRNAs in the bladder comes from the work done in BCa. Here we summarize the published data in order to identify the organ-specific miRNA expression patterns and draw comparisons between BCa and bladder dysfunction.
BCa is one of the most common cancers in the world [57], the fifth most commonly diagnosed non-cutaneous solid malignancy, and, after prostate cancer, the second most frequently diagnosed genitourinary tumor [58]. In 2008, 386,300 new cases of bladder cancers were diagnosed globally [59]. The early miRNA screening attempts were aimed at defining the cancer-specific miRNAs, and differentiating between the tumor stages by means of miRNA profiling. The first study in human bladder compared the miRNA expression profiles of bladder and kidney cancers [60]. Both types of cancers were found to share many miRNAs. The most significant up-regulated miRNAs in BCa versus normal bladder (1.2-fold change cutoff; p < 0.05) were: miR-223, miR-26b, miR-221, miR-103-1, miR-185, miR-23b, miR-203, miR-17-5p, miR-23a, and miR-205. However, this study did not identify any significantly down-regulated miRNA in tumors and the data did not overlap with the later published results [61], possibly because of the limited number of reference samples (two normal mucosa).
Expression of 343 miRNA was studied in noninvasive and invasive bladder carcinoma cell lines in order to identify the miRNA signature in BCa suitable for discriminating the superficial from the invasive disease [62]. Nine miRNAs with significant differential expression included mir-21, mir-31, mir-200a, mir-200c, mir-205, mir-373*, mir-487b, mir-498, and mir-503. MiR-21 expression was up-regulated and miR-205 down-regulated in the invasive compared with the noninvasive bladder cell lines. In 2009, Dyrskjot et al. investigated the expression profile of 290 microRNAs in immortalized urothelial cell lines, tumorigenic cell lines, 106 bladder tumors and 11 normal samples and identified some differentially expressed miRNAs between cancer and normal urothelium. In agreement with the previous study [62], miR-21 was the most up-regulated and miR-145 was the most down-regulated in cancer compared to the normal samples. Several differentially expressed miRNAs were found comparing different tumor stages. Most significant miRNAs up-regulated in progressing tumors were miR-129-5p, miR-518c*, miR-185, miR-133b, miR-373*, miR-320a, miR-145; and miRNAs, which were mostly down-regulated in progressing tumors were miR-29c, miR-29b, miR-29a, miR-361-5p, miR-203, and miR-205 [61].
Investigation of 322 miRNAs expressed in normal urothelium from patients with high-grade urothelial cell carcinomas (UCC) and disease-free controls revealed that 11 % of miRNAs were differentially expressed [63]. Down-regulation of miRNAs is a common phenomenon in low-grade tumors, and aberrant promoter hyper-methylation influences miRNA down-regulation in low-grade UCC. Many miRNAs down-regulated in low-grade tumors are predicted to target FGFR3 (miRs-99a/100/214/145/30a/125b/507). In contrast, in high-grade UCC up-regulation of many miRNAs including miRNA-21 can suppress p53 function [64].
Lin et al. performed miRNA profiling in BCa and matched normal urothelial epithelium controls and identified 37 up-regulated and 38 down-regulated miRNAs. Among them, miRNA-143 was most down-regulated, 13.7 times lower in tumor than in the matched control [65]. miRNA-143 was not expressed in the human BCa cell lines EJ and T24 and its transfection inhibited cell proliferation and reduced RAS protein levels [65]. Ichimi et al. investigated tumor-suppressive miRNAs in BCa [66]. Upon screening of 156 miRNAs in 14 BCas, five normal bladder epithelium (NBE) samples and three BCa cell lines, miR-145, miR-30a-3p, miR-133a, miR-133b, miR-195, miR-125b, and miR-199a* were shown to be significantly down-regulated in BCas [66]. Keratin 7 (KRT7) mRNA was a common predicted target for the down-regulated miRNAs. Compared to normal bladder epithelium, BCa samples had significantly higher mRNA levels of KRT 7 [66].
Using microarray technology, Song et al. reported that the expression profile of miRNAs was significantly altered in bladder urothelial carcinoma tissue compared to adjacent normal bladder tissue. Consistent with previous observations, most differentially expressed miRNAs were down-regulated. The top ten dis-regulated miRNAs including miR-1, miR-145, miR-143, miR-100, miR-200b, miR-708, miR-133a, miR-133b, and mir-125b were validated by real-time RT-PCR [67].
The first report of genome-wide miRNA expression profiling in human bladder urothelial carcinoma by deep sequencing was published in 2011 by Han et al. [68]. A total of 656 differentially expressed miRNAs were detected comprising known human miRNAs and miRNA antisense sequences (miRNA*s). MiR-490-5p was the most significantly down-regulated and miR-96 was the most significantly up-regulated miRNA [68].
Notwithstanding a long list of miRNAs de-regulated in BCa [69, 70], there is very little overlap in the patterns of miRNA expression between BCa and BPS (Table 1), which might be indicative of differential organ responses to specific diseases.
Inflammation in BPS and the role of miRNA in immune-mediated diseases
BPS is an inflammatory disorder
Inflammation occurs frequently in the lower urinary tract, and most often is a result of a urinary tract infection. One of the intriguing features of BPS is the presence of inflammation, often confirmed by biopsy, in the absence of an inflammatory agent (toxin or microorganism) [71]. Many causative factors have been suggested for BPS, including chronic or subclinical infection, autoimmunity and genetic susceptibility, which could be responsible for initiating the inflammatory response. However, a central role of inflammation has been confirmed in the pathogenesis of interstitial cystitis [72]. Epithelial dysfunction, often observed during BPS, might cause nerve sensitization, which in turn could lead to up-regulation of neurotransmitter release (tachykinins, glutamate, calcitonin gene-related peptide) [73]. Secretion of inflammatory mediators such as SP from sensory nerves has been implicated in the pathophysiology of pain triggering mast cell secretion/neurogenic inflammation [74]. Indeed, the obligatory requirement of tachykinin receptor NK1R in cystitis and its role in mast cell degranulation and inflammation have been confirmed in experiments with NK1R knockout mice [75]. Recently, increased urinary NGF levels were described in BPS/IC patients suggesting that chronic inflammation is involved in this bladder disorder [76].
A recent study investigated whether bladder inflammation could directly modulate the apoptotic signaling pathway in urothelial cells in interstitial cystitis/painful bladder syndrome (IC/PBS, equivalent definition of BPS) [77]. The levels of pro-apoptotic proteins, including phospho-p53 (Ser 15), Bad, Bax, and cleaved caspase-3 were significantly increased in the IC/PBS bladders. These results suggested that the tissue damage and abnormal urothelium in the IC/PBS bladders might be regulated concurrently by inflammatory signals, such as p38 mitogen-activated protein kinase and TNF-alpha. The contents of phospho-p38α and TNF-α in IC/PBS samples were significantly greater than in the control. The in vitro analysis showed that the apoptotic process could be induced by TNF-alpha treatment and anisomycin stimulation in normal urothelial cells [77].
miRNAs as regulators of immune response
During the recent years, plentiful evidence has been amassed pointing to the critical role of miRNAs both in the development of immune system and its function in innate and adaptive responses [78]. Several miRNAs, including miR-155, miR-181a, miR-146a, miR-150, miR-223 and miR-17-92 have been shown to regulate development, differentiation, and function of immune cells (reviewed by [79, 80]). The innate immune response is a cellular response, comprising macrophages, monocytes, granulocytes, and dendritic cells, which is started by activation of Toll-like receptors (TLR). TLR signaling induces numerous miRNAs including miR-155, miR-146a, and miR-21 [81]. Additionally, miR-125b and let-7 are important in controlling innate immune responses: miR-125b targets TNF-α, while let-7 targets IL-6 mRNA [82, 83]. Interestingly, although in our study we detected an increased neutrophil infiltration in the BPS patients [13], none of the classical immune cell-specific miRNAs were significantly up-regulated. However, we detected an increase in miR-500* and miR-511, which are important for the dendritic cell, monocyte and macrophage function [84, 85]. These results indicate that the elevated miRNAs levels might originate from the bladder tissue remodeling during BPS, rather than the immune cell infiltration.
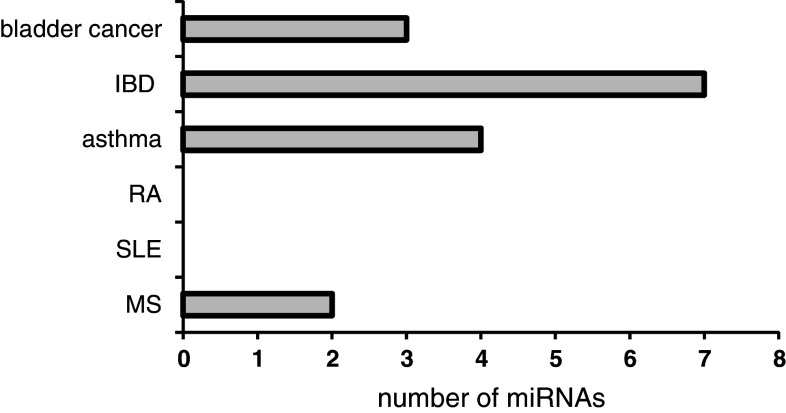
Taking into account the role of inflammation in BPS and the lack of data on miRNA regulation in the similar inflammatory disorders of the bladder, we compared the miRNAs identified in other immune-mediated diseases, to the BPS profile. These data are summarized in Table 1 and Fig. 2.
Fig. 2.
Similarly regulated miRNAs. Number of miRNAs either up- or down-regulated both in BPS and BCa, IBD, RA, SLE, or MS
Asthma
Asthma is a common chronic airway disease characterized by airway remodeling (epithelial alteration, fibrosis, smooth muscle hypertrophy) and associated with Th2 response that stimulates eosinophile and mast cell infiltration. Asthma is a common co-morbidity of BPS [86], therefore we correlated the available information on miRNA profiling in this disorder with our data in BPS [13]. In the mouse models of asthma, miR-1 was down- and miR-21 up-regulated, along with 21 other differentially expressed miRNAs, and in another model, miR-16, -21, and -126 were up-regulated. MiR-133, -25, 146a- and -26a regulate human airway smooth muscle cells in asthma models (reviewed by [87]). MiR-23a, -23b, and -25 play an important role in regulating the phenotype of airway smooth muscle via modulating the expression of inflammatory mediators like RANTES, eotaxin, and TNF-α. These genes are responsible for extracellular matrix turnover and expression of contractile proteins (myosin heavy chain) [88]. Interestingly, some of the miRNAs described in this study appear to be important in BPS—we found miR-23a, -23b, -25, and -320 up-regulated in BPS, arguing for an increased smooth muscle phenotype, which is consistent with low bladder volumes and thick bladder walls observed in patients [13].
Autoimmune diseases
The role of miRNAs in autoimmune diseases including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and multiple sclerosis (MS) has been postulated and confirmed by altered levels of several important miRNAs (reviewed in [79]). Although no data support a direct causal role of autoimmune reactivity in the pathogenesis of BPS/IC, there is ample indirect evidence, such as the strong female preponderance and the clinical association between BPS and other known autoimmune diseases within patients and families [15]. BPS is associated with the diagnosis of RA [89], SLE, and Sjögren’s syndrome [90]. In RA, miRNAs miR-146a, miR-223, and miR-155, characteristic of immune cell activation, were significantly up-regulated in synovial fluid samples [91]. In MS, miRNAs miR-34a, -155 and -326 were elevated in active lesions [92]. In SLE, miR-146a, a negative regulator of TLR signaling was profoundly decreased, whereas miR-148a and miR-21 were increased in T cells from patients with lupus [93]. Both miR-422a and miR-22 have previously been implicated in MS [94]. Taken together, miRNA profiling in autoimmune diseases revealed a strong prevalence of immune cell-specific miRNAs and some overlap with BPS profile established in our study [13].
Inflammatory bowel disease (IBD)
IBD, comprising Crohn’s disease (CD) and ulcerative colitis (UC), is a gastrointestinal chronic inflammatory disorder and a frequent co-morbidity of BPS [95]. Earlier work has demonstrated that both the number of SP-positive nerve endings and mast cell count were increased in patients with BPS and IBD [96], implicating a similar pathogenic mechanism. There are numerous reports indicating that patients with CD and UC have altered miRNA profiles [97–105]. Analysis of miRNA expression in patients with active UC, inactive UC, CD, irritable bowel syndrome, infectious colitis, and microscopic colitis revealed eight miRNAs (miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195, and Let-7f) which were significantly increased in active UC tissues and three miRNAs (miR-192, miR-375, and miR-422b) were significantly decreased in the UC tissues. Up-regulation of miR-192 was observed in IBD-associated dysplasia [104]. UC patients show an up-regulation of immune cell-specific miR-21 and miR-155 in inflamed tissue [106].
Comparing active to quiescent UC, it was reported that four miRNAs (miR-188-5p, miR-25, miR-320a, miR-346) were down-regulated and five miRNA (miR-29a, miR-29b, miR-126*, miR-127-3p, miR-324-3p) were up-regulated [103]. We found a strong overlap in miRNA expression profiles between BPS and IBD (Table 1).
Inflammatory pain disorders
Pain is one of the hallmarks of inflammation, and chronic inflammatory conditions are often accompanied by chronic pain. Inflammatory pain serves as a warning that hinders the normal bodily function until the stimulus abates or the tissue repairs. Noxious stimuli activate numerous receptors and ion channels, and signals propagate to the central nervous system, where pain is perceived [107]. Epigenetic modifications influence inflammatory cytokine metabolism, steroid responsiveness, and opioid sensitivity thus contributing to the development of chronic pain [108], however, there are little data implicating miRNA in peripheral pain. It was shown that miRNAs might regulate the action of opioid drugs: let-7 miRNA family, including let-7 g, which is up-regulated in BPS, was found to be critical for μ opioid receptor function. Chronic exposure to morphine caused the reduction of MOR levels and concomitant increase in let-7 miRNA synthesis [109]. It is possible that miRNA induction might serve as an adaptive response, aimed at reducing pain and inflammation: the animals with neuropathic pain showed significant improvements after infusion of miR-23b. This miRNA down-regulated NADPH oxidase 4 (NOX4), a reactive oxygen species (ROS) family member overexpressed in neuropathic pain [110].
MiR-124a is expressed in the spinal cord and has been implicated in pain. Knock-down of miRNA-124a increased the nociceptive behavior via an up-regulation of its pain-relevant target MeCP2 and proinflammatory marker genes [111]. Another recent study identified miR-181a as a modulator of GABA(Aα-1) receptor subunit down-regulation in spinal cord following neonatal cystitis-induced chronic visceral pain in rats [112].
Several miRNAs were profiled from whole blood samples of patients with complex regional pain syndrome (CRPS), a chronic pain condition resulting from dysfunction of central or peripheral nervous systems [113]. Like BPS, CRPS bears many signs of neurogenic inflammation. MiRNA profiling identified differential expression of 18 miRNAs in CRPS patients. Most miRNAs were down-regulated, however there is a significant overlap between the miRNAs, altered in CRPS and BPS (Table 1).
Functional information on miRNAs, altered in BPS
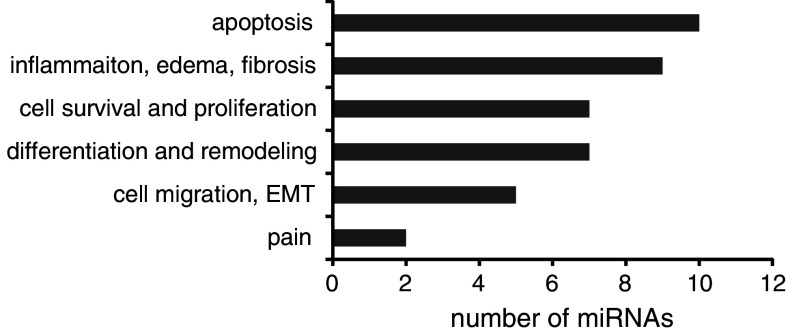
At the time of miRNA profiling in BPS, very few miRNA targets were validated, making it difficult to determine their function and potential importance for the disease pathogenesis. Since then, there has been a surge of publications on the miRNA-regulated proteins in cell lines and disease states. Sometimes multiple targets have been attributed to the same miRNA, implicating it in several, often contradictory, cellular functions. This divergence reflects the pleiotropic activity of miRNAs, which are known to regulate multiple mRNAs [1, 9]. Nevertheless, out of 31 miRNAs from our original study, no targets have been defined for miR-30e-5p, miR-342-3p, miR-485-5p, miR-502-5p, miR-572, miR-594, and miR-597. In contrast, numerous proteins have been identified as effectors of miRNAs belonging to the well-characterized families like let-7. The information on the validated targets of miRNAs altered in BPS is summarized in Table 2 and Fig. 3. Below, we refer in more detail to the functional data, relevant for BPS pathogenesis.
Table 2.
Validated BPS miRNA targets
| miRNA | Regulation in BPS | Target; function | Reference |
|---|---|---|---|
| let-7 g | Up |
1. Lectin-like oxidized low-density lipoprotein receptor-1; inhibition of SMC apoptosis 2. Type I collagen alpha2 (COL1A2); inhibition of migration 3. μ opioid receptor; role in pain |
1. [115] 2. [116] 3. [109] |
| miR-23a | Up |
1. Transcription factor Foxo3a; role in cardiac hypertrophy 2. Muscle-specific ubiquitin ligases, MAFbx/atrogin-1 and muscle RING-finger 1 (MuRF1); inhibition of muscle atrophy 3. Sprouty2 and Sema6A proteins; promote angiogenic activity. 4. E-cadherin |
1. [117] 2. [118] 3. [154] 4. [119] |
| miR-23b | Up |
1. Smads 3, 4 5; regulation of TGF-β signaling 2. FZD7 or MAP3k1; metastasis control 3. TAB 2, TAB 3, and IKK-α; suppresses IL-17-induced inflammation |
1. [121] 2. [120] 3. [122] |
| miR-25 | Up |
1. Bim, pro-apoptotic protein; increase in proliferation, reduction of apoptosis 2. TRAIL Death Receptor-4; reduction of apoptosis |
1. [124] 2. [125] |
| miR-26a | Up |
1. SMAD1, 4; increase SMC proliferation, delay differentiation 2. Glycogen synthase kinase GSK-3β; smooth muscle hypertrophy 3. Cyclin D2, EZH2; inhibition of cancer cell proliferation |
1. [126] 2. [127] 3. [155] |
| miR-27a | Up |
1. Sprouty2 and Sema6A; enhance angiogenesis 2. Myostatin; role in skeletal muscle proliferation 3. Sprouty 2; enhances proliferation |
1. [154] 2. [123] 3 [156]. |
| miR-30e-5p | Up | No validated targets | |
| miR-95 | Up | Sorting nexin 1 (SNX1); increases proliferation in cancer cells | [129] |
| miR-130b | Up |
ZEB1; represses EMT in cancer cells 2. RUNX3 apoptotic protein; increase cell proliferation |
1. [130] 2 [157]. |
| miR-133b | Up |
1. Pitx3 transcription factor; role in neuron differentiation and pain 2. FAIM, GSTP; promotes apoptosis |
1. [152] 2. [158] |
| miR-148a | Down |
1. ROCK1; increase motility 2. WNT1 and WNT10B; activation of WNT/β-catenin pathway |
1. [131] 2. [133] |
| miR-182 | Down |
1. FOXO1; proliferation 2. CREB1; inhibition of proliferation |
1. [159] 2. [160] |
| miR-186 | Up | P2X7; Ca homeostasis | [134] |
| miR-192 | Up |
1. SIP1, Ebox repressor; increase of Col1a2 and fibrosis 2. ZEB1/2; increase in fibrosis |
1. [135] 2. [136] |
| miR-199a(-5p) | Up |
1. Cell junction proteins LIN7C, ARHGAP12, PALS1, RND1 and PVRL1; causes epithelial dysfunction 2. HIF1alpha, Sirtuin; hypertrophy in cardiomyocytes 3. Discoidin domain receptor 1; decrease cancer cell proliferation |
1. [14] 2. [161] 3. [162] |
| miR-320(a) | Up |
1. β-Catenin; suppresses cancer cell proliferation 2. Neuropilin 1; suppression of cell proliferation 3. Aquaporins 1 and 4; increase of edema 4. NK1R; role in neurogenic inflammation |
1. [139] 2. [140] 3. [141] 4. [13] |
| miR-324-3p | Up | Prolyl endopeptidase; promotes fibrosis | [142] |
| miR-328 | Up |
1. NK1R; role in neurogenic inflammation 2. L-type calcium channel a1C, IGFR1; smooth muscle remodeling |
1. [13] 2. [143] |
| miR-342(-3p) | Up | No functional data | |
| miR-379 | Up | ABCC2 transporter; inhibition of efflux for various endogenous and exogenous compounds | [163] |
| miR-422a | Up | Tumor suppressor | [164] |
| miR-422b(378) | Up |
1. IGFR1; cardiac remodeling and decreased survival 2. Sufu (Suppressor of fused) and Fus-1; increased cell survival and proliferation |
1. [144] 2. [165] |
| miR-449b | Up |
1. NK1R; role in neurogenic inflammation 2. CDK6 and CDC25A; cell cycle arrest |
1. [13] 2. [145] |
| miR-485-5p | Up | No functional data | |
| miR-493 | Down |
1. FZD4 and RhoC; decrease bladder cancer cell growth and migration 2. IGF1R; inhibition of cell growth and metastasis |
1. [146] 2. [147] |
| miR-500(*) | Up |
1. NK1R; role in neurogenic inflammation 2. Marker of dendritic cells and monocytes, regulation of immune response |
1 [13]. 2. [85] |
| miR-502(-5p) | Up | No functional data | |
| miR-511 | Up | TLR4 and CD80; regulation of immune response | [84] |
| miR-572 | Up | No functional data | |
| miR-594 | Up | No functional data | |
| miR-597 | Up | No functional data |
Fig. 3.
miRNA functions. Number of miRNAs, altered in BPS and associated with a specific function
Let-7g belongs to the Let-7 family of miRNAs. Let-7 miRNAs regulate cell differentiation, proliferation, and neuromuscular development [16, 114]. The specific functions of let-7g include the inhibition of smooth muscle cell (SMC) apoptosis by targeting lectin-like oxidized low-density lipoprotein receptor-1 [115]. Additionally, it targets type I collagen alpha2 (COL1A2) [116], as well as μ opioid receptor, and is predicted to have a role in pain perception [109].
miR-23a, -23b, and -27 are members of miR-23 ∼ 27 ∼ 24 gene cluster. miR-23a is a versatile regulator of muscle development and promotes cardiac hypertrophy [117]. Ectopic expression of miR-23a was sufficient to protect muscles from atrophy in vitro and in vivo [118]. In cancer cell lines, the expression of miR-23a resulted in inhibition of E-cadherin expression [119]. Interestingly, we and the others have shown a decrease of E-cadherin levels in BPS, concomitant with an up-regulation of miR-23a. MiR-23b has pleiotropic functions in cell proliferation, and has been altered in many cancers [120]. It targets and down-regulates Smads and consequently TGF-beta signaling [121]. Recently, an important function of this miRNAs in regulation of autoimmune responses has been highlighted: miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TGF-β-activated kinase 1/MAP3K7 binding protein 2 (TAB 2), TAB 3, and inhibitor of nuclear factor κ-B kinase subunit α (IKK-α) causing a decrease of IL-17-, tumor necrosis factor α (TNF-α)- or IL-1β-induced NF-κB activation and inflammatory cytokine expression and repression of autoimmune inflammation[122]. miR-27 plays a role in cell survival, and its overexpression in C2C12 cells resulted in myoblast proliferation by reducing the expression of myostatin, a critical inhibitor of skeletal myogenesis [123].
miR-25 was found up-regulated in BPS and in a mouse model of OAB, where it has been suggested to down-regulate P2X receptors, although the authors did not proceed beyond the in silico analysis [56]. miR-25 stimulates cell proliferation in ovarian and other cancers [124]. It was shown to protect cells against TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis [125].
miR-26a is an important regulator of smooth muscle proliferation and function. miR-26a promotes vascular SMC proliferation while inhibiting cellular differentiation and apoptosis, and alters TGF-β pathway signaling [126]. Overexpression of miRNA-26a blunted SMC differentiation. miRNA-26a influences TGF-β-pathway signaling by targeting Smad-1 and Smad-4. Mechanical stretch up-regulates miR-26a expression and consequently induces SMC proliferation leading to hypertrophy [127]. miR-26a is a regulator of skeletal muscle: it was induced during skeletal muscle regeneration and its inhibition de-repressed Smad activity and inhibited differentiation [128].
miR-95 expression is up-regulated in many tumors. Mechanistic studies revealed that miR-95 repressed the expression of sorting nexin 1 (SNX1), whereas miR-95 silencing up-regulated SNX1 expression [129].
miR-130b targets and inhibits transcription factor ZEB1, suppressing EMT transition. Its transcription depends on p53, and repressed expression of miR-130b triggered ZEB1-dependent EMT [130].
miR-148a is down-regulated in BPS. Rho-associated coiled-coil containing protein kinase 1 (ROCK1) is one of its main targets. In skeletal muscle, increase of miR-148a helps myogenic differentiation through inhibition of ROCK1 [131], and in cancer cells it leads to suppressed tumor cell invasion and metastasis [132]. Similarly, silencing miR-148a in cancerous fibroblasts stimulated cell motility by de-repressing its targets WNT1 and WNT10B and activation of WNT/β-catenin pathway [133].
miR-186 was shown to target P2X7 Ca channel, implicated in cell survival and apoptosis [134].
miR-192 is highly expressed in the kidney epithelial cells, where it controls TGF-β-induced collagen 1 a2 expression via down-regulating Smad-interacting protein SIP1, a E-box repressor [135]. In a separate study, it was shown to increase collagen expression by targeting the E-box repressors Zeb1/2 [136]. Both studies allow concluding that miR-192 promotes fibrosis.
miR-199a-5p is an important regulator of intercellular junctions. Upon overexpression in urothelial cells, it impairs correct tight junction formation and leads to increased permeability. MiR-199a-5p directly targets mRNAs encoding LIN7C, ARHGAP12, PALS1, RND1 and PVRL1 and attenuates their expression levels to a similar extent. It is predominantly expressed in the bladder smooth muscle, but also detected in the mature bladder urothelium and primary urothelial cultures [14].
MiR-199a-5p is up-regulated in cardiac hypertrophy and its overexpression in cardiomyocytes leads to cell-size increase [137]. A separate study claims that in cultured cell lines, the expression of miR-199a and miR-199a* (miR-199a/a*) is confined to fibroblasts [138].
miR-320a has anti-proliferative effect, which it exerts by targeting β-catenin [139], neuropilin 1 (NRP-1), which is a co-receptor of vascular epithelial growth factor [140], and aquaporins 1 and 4 [141]. We have shown that miR-320a down-regulates tachykinin NK1 receptor [13].
miR-324-3p is implicated in the pathogenesis of renal fibrosis. A predicted target of miR-324-3p is prolyl endopeptidase. In cultured tubular cells, transient transfection with a miR-324-3p mimic increased deposition of collagen [142].
miR-328 is regulating the NK1R expression levels [13], and also has a role in smooth muscle remodeling by targeting L-type calcium channel-alpha1C and the insulin growth factor 1 receptor expression, ultimately leading to apoptosis of pulmonary arterial smooth muscle cells [143].
miR-378 is a cardioabundant microRNA that targets IGF1R. In tissues such as fibroblasts and fetal hearts, where IGF1 levels are high, there were either absent or significantly low miR-378 levels, suggesting an inverse relationship between these two factors [144]. miR-449b, in addition to influencing NK1R expression, targets and inhibits oncogenic CDK6 and CDC25A, resulting in cell-cycle arrest [145].
miR-493 is down-regulated in BPS. Interestingly, it has been implicated in regulation of bladder cancer tumorigenesis: expression of miR-493 in bladder cancer (T24, J82, and TCCSUP) cells and tissues was down-regulated. miR-493 decreased cell growth and migration by reducing the protein expression of FZD4 and RhoC [146]. IGF1R was identified as a direct target of miR-493, and its inhibition partially mimicked the anti-metastatic effects [147].
miR-550* was identified by us as a regulator of NK1R expression. Recently, it was shown to be specific for plasmacytoid dendritic cells (pDC) and monocytes [85]. miR-511 is an important regulator of immune cell function in dendritic cells and macrophages: inhibition of the two most highly up-regulated miRNAs, miR-511 and miR-99b, resulted in reduced lower DC-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) level. Prediction of miRNA-511 targets revealed a number of genes with known immune functions, of which TLR4 and CD80 were validated. Interestingly, under the cell-cycle arrest conditions, miR-511 seems to function as a positive regulator of TLR4 [84].
Conclusions and future directions
Here we conducted a comparative analysis of the miRNA expression profile in BPS, BCa, and several inflammatory disorders, and summarized the validated functional information on the differentially expressed miRNAs. Despite the existence of several miRNA profiling studies in BCa, we found strikingly little overlap between the similarly regulated miRNA species in BCa and BPS.
On the other hand, seven out of 31 miRNA, altered in BPS showed the same regulatory pattern in IBD (Table 1, Fig. 2). IBD shares many features with BPS, including inflammation, pain, smooth muscle remodeling, and changes in epithelial permeability [97]. Disruption of epithelial barrier function was identified as one of the pathologic mechanisms in IBD, and miR-199a-5p, which we have recently characterized as a major regulator of urothelial tight and adherens junctions [14], is also up-regulated in IBD patients (Table 1). Similarly, fibrosis is a common feature of an advanced inflammatory disease and has been described in both IBD [148] and BPS [149]. Interestingly, miR-192, up-regulated in both disorders (Table 1), has validated targets whose repression leads to increased fibrosis (Table 2). Based on this information, it would be interesting to take a closer look at these miRNAs in order to determine their therapeutic potential.
We found less overlap between BPS miRNAs and miRNAs de-regulated in the other inflammatory and autoimmune diseases, with the exception of asthma, which shared four up-regulated miRNAs. All of them (miR-23a, -23b, -25, and -26a) have been implicated in smooth muscle remodeling, and some have validated targets among the proteins involved in the regulation of muscle growth and differentiation (Table 2). The majority of BPS patients enrolled in our study had low-volume thick-walled bladders [13], and it would be tempting to speculate on the role of these four elevated miRNA in SMC proliferation during BPS.
Some of the functions of the miRNAs altered in BPS could be deduced based on the information about their validated protein targets identified in other cell systems (Fig. 3). Due to the pleiotropic nature of miRNAs, the same miRNA is often implicated in the regulation of opposing processes, i.e., promoting both cell proliferation and apoptosis. Based on the data in Table 2, we grouped the miRNAs according to their functions (Fig. 3). Most of miRNAs, altered in BPS (ten out of 31) have validated targets whose down-regulation results in increased apoptosis. These findings fit well with the emerging data demonstrating increased apoptosis in BPS [77]. The second prominent group was miRNAs influencing inflammation, edema, and fibrosis (nine out of 31). We also identified several miRNAs whose targets include regulators of cell proliferation and differentiation. It would be interesting to determine which bladder layers harbors the miRNAs belonging to these functionally opposing groups, in order to evaluate the differential effects of inflammation on the bladder urothelium and smooth muscle.
MicroRNA research is an exciting and challenging field, and we are witness to its burgeoning. Recent years have brought about a flood of publications containing both the expression profiling and target validation data. Bringing these results together yields helpful insights into the miRNA function in human diseases and their potential therapeutic applications.
Acknowledgments
We gratefully acknowledge the financial support of the Swiss National Science Foundation (SNF Grant 320030_135783/1 to K. Monastyrskaya).
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27(24):3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 7.Huang S, Wu S, Ding J, Lin J, Wei L, Gu J, He X. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 2010;38(20):7211–7218. doi: 10.1093/nar/gkq564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39(16):6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS ONE. 2008;3(10):e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilmott JS, Zhang XD, Hersey P, Scolyer RA. The emerging important role of microRNAs in the pathogenesis, diagnosis and treatment of human cancers. Pathology. 2011;43(6):657–671. doi: 10.1097/PAT.0b013e32834a7358. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez Freire V, Burkhard FC, Kessler TM, Kuhn A, Draeger A, Monastyrskaya K. MicroRNAs may mediate the down-regulation of neurokinin-1 receptor in chronic bladder pain syndrome. Am J Pathol. 2010;176(1):288–303. doi: 10.2353/ajpath.2010.090552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monastyrskaya K, Sanchez-Freire V, Gheinani AH, Klumpp DJ, Babiychuk EB, Draeger A, Burkhard FC. miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. Am J Pathol. 2012 doi: 10.1016/j.ajpath.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 15.van de Merwe JP. Interstitial cystitis and systemic autoimmune diseases. Nat Clin Pract Urol. 2007;4(9):484–491. doi: 10.1038/ncpuro0874. [DOI] [PubMed] [Google Scholar]
- 16.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 17.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 18.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Nat Acad Sci USA. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Nat Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Nat Acad Sci USA. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204(7):1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. NeuroReport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2(7):e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16(12):939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 27.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 29.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Xu X, Ma Z, Huo Y, Xiao Z, Li Y, Wang Y. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA. 2011;17(8):1511–1528. doi: 10.1261/rna.2732611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14(10):934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 37.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(suppl 1):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Merwe JP, Nordling J, Bouchelouche P, Bouchelouche K, Cervigni M, Daha LK, Elneil S, Fall M, Hohlbrugger G, Irwin P, Mortensen S, van Ophoven A, Osborne JL, Peeker R, Richter B, Riedl C, Sairanen J, Tinzl M, Wyndaele JJ. Diagnostic criteria, classification, and nomenclature for painful bladder syndrome/interstitial cystitis: an ESSIC proposal. Eur Urol. 2008;53(1):60–67. doi: 10.1016/j.eururo.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Curhan GC, Speizer FE, Hunter DJ, Curhan SG, Stampfer MJ. Epidemiology of interstitial cystitis: a population based study. J Urol. 1999;161(2):549–552. [PubMed] [Google Scholar]
- 45.Fall M, Oberpenning F, Peeker R. Treatment of bladder pain syndrome/interstitial cystitis 2008: can we make evidence-based decisions? Eur Urol. 2008;54(1):65–75. doi: 10.1016/j.eururo.2008.03.086. [DOI] [PubMed] [Google Scholar]
- 46.Lilly JD, Parsons CL. Bladder surface glycosaminoglycans is a human epithelial permeability barrier. Surg Gynecol Obstet. 1990;171(6):493–496. [PubMed] [Google Scholar]
- 47.Moskowitz MO, Byrne DS, Callahan HJ, Parsons CL, Valderrama E, Moldwin RM. Decreased expression of a glycoprotein component of bladder surface mucin (GP1) in interstitial cystitis. J Urol. 1994;151(2):343–345. doi: 10.1016/s0022-5347(17)34944-3. [DOI] [PubMed] [Google Scholar]
- 48.Birder LA, de Groat WC. Mechanisms of disease: involvement of the urothelium in bladder dysfunction. Nat Clin Pract Urol. 2007;4(1):46–54. doi: 10.1038/ncpuro0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons CL. The role of the urinary epithelium in the pathogenesis of interstitial cystitis/prostatitis/urethritis. Urology. 2007;69(4 Suppl):9–16. doi: 10.1016/j.urology.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 50.Tomaszewski JE, Landis JR, Russack V, Williams TM, Wang LP, Hardy C, Brensinger C, Matthews YL, Abele ST, Kusek JW, Nyberg LM. Biopsy features are associated with primary symptoms in interstitial cystitis: results from the interstitial cystitis database study. Urology. 2001;57(6 Suppl 1):67–81. doi: 10.1016/s0090-4295(01)01166-9. [DOI] [PubMed] [Google Scholar]
- 51.Slobodov G, Feloney M, Gran C, Kyker KD, Hurst RE, Culkin DJ. Abnormal expression of molecular markers for bladder impermeability and differentiation in the urothelium of patients with interstitial cystitis. J Urol. 2004;171(4):1554–1558. doi: 10.1097/01.ju.0000118938.09119.a5. [DOI] [PubMed] [Google Scholar]
- 52.Zhang CO, Wang JY, Koch KR, Keay S. Regulation of tight junction proteins and bladder epithelial paracellular permeability by an antiproliferative factor from patients with interstitial cystitis. J Urol. 2005;174(6):2382–2387. doi: 10.1097/01.ju.0000180417.11976.99. [DOI] [PubMed] [Google Scholar]
- 53.Liu HT, Shie JH, Chen SH, Wang YS, Kuo HC. Differences in mast cell infiltration, E-cadherin, and zonula occludens-1 expression between patients with overactive bladder and interstitial cystitis/bladder pain syndrome. Urology. 2012;80:225. doi: 10.1016/j.urology.2012.01.047. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Freire V, Blanchard MG, Burkhard FC, Kessler TM, Kellenberger S, Monastyrskaya K. Acid-sensing channels in human bladder: expression, function and alterations during bladder pain syndrome. J Urol. 2011;186(4):1509–1516. doi: 10.1016/j.juro.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 55.Sadegh MK, Ekman M, Rippe C, Uvelius B, Sward K, Albinsson S. Deletion of Dicer in smooth muscle affects voiding pattern and reduces detrusor contractility and neuroeffector transmission. PLoS ONE. 2012;7(4):e35882. doi: 10.1371/journal.pone.0035882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S, Lv JW, Yang P, Yu Q, Pang J, Wang Z, Guo H, Liu S, Hu J, Li J, Leng J, Huang Y, Ye Z, Wang CY. Loss of dicer exacerbates cyclophosphamide-induced bladder overactivity by enhancing purinergic signaling. Am J Pathol. 2012;181(3):937–946. doi: 10.1016/j.ajpath.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 57.Zhu J, Jiang Z, Gao F, Hu X, Zhou L, Chen J, Luo H, Sun J, Wu S, Han Y, Yin G, Chen M, Han Z, Li X, Huang Y, Zhang W, Zhou F, Chen T, Fa P, Wang Y, Sun L, Leng H, Sun F, Liu Y, Ye M, Yang H, Cai Z, Gui Y, Zhang X. A systematic analysis on dna methylation and the expression of both mRNA and microRNA in bladder cancer. PLoS ONE. 2011;6(11):e28223. doi: 10.1371/journal.pone.0028223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theodorescu D. Molecular pathogenesis of urothelial bladder cancer. Histol Histopathol. 2003;18(1):259–274. doi: 10.14670/HH-18.259. [DOI] [PubMed] [Google Scholar]
- 59.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 60.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25(5):387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 61.Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, Kauppinen S, Ulhoi BP, Kjems J, Borre M, Orntoft TF. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69(11):4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 62.Neely LA, Rieger-Christ KM, Neto BS, Eroshkin A, Garver J, Patel S, Phung NA, McLaughlin S, Libertino JA, Whitney D, Summerhayes IC. A microRNA expression ratio defining the invasive phenotype in bladder tumors. Urol Oncol. 2008;28(1):39–48. doi: 10.1016/j.urolonc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 63.Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, Larre S, Milo M, Rehman I, Rosario DJ, Di Martino E, Knowles MA, Meuth M, Harris AL, Hamdy FC. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69(21):8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayala de la Pena F, Kanasaki K, Kanasaki M, Tangirala N, Maeda G, Kalluri R. Loss of p53 and acquisition of angiogenic microRNA profile are insufficient to facilitate progression of bladder urothelial carcinoma in situ to invasive carcinoma. J Biol Chem. 2011;286(23):20778–20787. doi: 10.1074/jbc.M110.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181(3):1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 66.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125(2):345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 67.Song T, Xia W, Shao N, Zhang X, Wang C, Wu Y, Dong J, Cai W, Li H. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010;11(4):905–911. [PubMed] [Google Scholar]
- 68.Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, Jiang Z, Zhang Z, Yang R, Li Z, Tang A, Li X, Ye J, Guan Z, Gui Y, Cai Z. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS ONE. 2011;6(3):e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59(5):671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 70.Ru Y, Dancik GM, Theodorescu D. Biomarkers for prognosis and treatment selection in advanced bladder cancer patients. Curr Opin Urol. 2011;21(5):420–427. doi: 10.1097/MOU.0b013e32834956d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wyndaele JJ, Van Dyck J, Toussaint N. Cystoscopy and bladder biopsies in patients with bladder pain syndrome carried out following ESSIC guidelines. Scand J Urol Nephrol. 2009;43(6):471–475. doi: 10.3109/00365590903199007. [DOI] [PubMed] [Google Scholar]
- 72.Grover S, Srivastava A, Lee R, Tewari AK, Te AE. Role of inflammation in bladder function and interstitial cystitis. Ther Adv Urol. 2011;3(1):19–33. doi: 10.1177/1756287211398255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keay S. Cell signaling in interstitial cystitis/painful bladder syndrome. Cell Signal. 2008;20(12):2174–2179. doi: 10.1016/j.cellsig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 74.Pang X, Marchand J, Sant GR, Kream RM, Theoharides TC. Increased number of substance P positive nerve fibres in interstitial cystitis. Br J Urol. 1995;75(6):744–750. doi: 10.1111/j.1464-410x.1995.tb07384.x. [DOI] [PubMed] [Google Scholar]
- 75.Saban R, Saban MR, Nguyen NB, Lu B, Gerard C, Gerard NP, Hammond TG. Neurokinin-1 (NK-1) receptor is required in antigen-induced cystitis. Am J Pathol. 2000;156(3):775–780. doi: 10.1016/S0002-9440(10)64944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu HT, Kuo HC. Increased urine and serum nerve growth factor levels in interstitial cystitis suggest chronic inflammation is involved in the pathogenesis of disease. PLoS ONE. 2012;7(9):e44687. doi: 10.1371/journal.pone.0044687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shie JH, Liu HT, Kuo HC (2012) Increased cell apoptosis of urothelium mediated by inflammation in interstitial cystitis/painful bladder syndrome. Urology 79 (2):484 e487-413. doi:10.1016/j.urology.2011.09.049 [DOI] [PubMed]
- 78.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 79.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157(4):163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tomankova T, Petrek M, Gallo J, Kriegova E. MicroRNAs: emerging regulators of immune-mediated diseases. Scand J Immunol. 2011 doi: 10.1111/j.1365-3083.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 81.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11(3):163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 82.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 84.Tserel L, Runnel T, Kisand K, Pihlap M, Bakhoff L, Kolde R, Peterson H, Vilo J, Peterson P, Rebane A. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J Biol Chem. 2011;286(30):26487–26495. doi: 10.1074/jbc.M110.213561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D’Asaro M, Chiappe A, Sridhar S, Pacheco GD, Burczynski ME, Hochstrasser D, Vonderscher J, Matthes T. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS ONE. 2012;7(1):e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keller JJ, Chen YK, Lin HC. Comorbidities of bladder pain syndrome/interstitial cystitis: a population-based study. BJU Int. 2012 doi: 10.1111/j.1464-410X.2012.11539.x. [DOI] [PubMed] [Google Scholar]
- 87.Jiang X. The emerging role of microRNAs in asthma. Mol Cell Biochem. 2011;353(1–2):35–40. doi: 10.1007/s11010-011-0771-z. [DOI] [PubMed] [Google Scholar]
- 88.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Resp Cell Mol Biol. 2010;42(4):506–513. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keller JJ, Liu SP, Lin HC. A case-control study on the association between rheumatoid arthritis and bladder pain syndrome/interstitial cystitis. Neurourol Urodyn. 2012 doi: 10.1002/nau.22348. [DOI] [PubMed] [Google Scholar]
- 90.van de Merwe JP, Yamada T, Sakamoto Y. Systemic aspects of interstitial cystitis, immunology and linkage with autoimmune disorders. Int J Urol. 2003;10(Suppl):S35–S38. doi: 10.1046/j.1442-2042.10.s1.10.x. [DOI] [PubMed] [Google Scholar]
- 91.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 93.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 94.Siegel SR, Mackenzie J, Chaplin G, Jablonski NG, Griffiths L. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep. 2012;39(5):6219–6225. doi: 10.1007/s11033-011-1441-7. [DOI] [PubMed] [Google Scholar]
- 95.Chelimsky G, Heller E, Buffington CA, Rackley R, Zhang D, Chelimsky T. Co-morbidities of interstitial cystitis. Front Neurosci. 2012;6:114. doi: 10.3389/fnins.2012.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pang X, Boucher W, Triadafilopoulos G, Sant GR, Theoharides TC. Mast cell and substance P-positive nerve involvement in a patient with both irritable bowel syndrome and interstitial cystitis. Urology. 1996;47(3):436–438. doi: 10.1016/S0090-4295(99)80469-5. [DOI] [PubMed] [Google Scholar]
- 97.Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18(1):187–193. doi: 10.1002/ibd.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iborra M, Bernuzzi F, Invernizzi P, Danese S. MicroRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. Autoimmun Rev. 2012;11(5):305–314. doi: 10.1016/j.autrev.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 99.Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012 doi: 10.1016/j.crohns.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 100.Zwiers A, Kraal L, van de Pouw Kraan TC, Wurdinger T, Bouma G, Kraal G. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol. 2012;188(4):1573–1577. doi: 10.4049/jimmunol.1101494. [DOI] [PubMed] [Google Scholar]
- 101.Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE, Crawford NP, Galandiuk S. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat. 2012;33(3):551–560. doi: 10.1002/humu.22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dalal SR, Kwon JH. The role of microRNA in inflammatory bowel disease. Gastroenterol Hepatol (NY) 2010;6(11):714–722. [PMC free article] [PubMed] [Google Scholar]
- 103.Fasseu M, Treton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soule JC, Moreau R, Bouhnik Y, Laburthe M, Groyer A, Ogier-Denis E (2010) Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One 5 (10). doi:10.1371/journal.pone.0013160 [DOI] [PMC free article] [PubMed]
- 104.Olaru AV, Selaru FM, Mori Y, Vazquez C, David S, Paun B, Cheng Y, Jin Z, Yang J, Agarwal R, Abraham JM, Dassopoulos T, Harris M, Bayless TM, Kwon J, Harpaz N, Livak F, Meltzer SJ. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis. 2011;17(1):221–231. doi: 10.1002/ibd.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clark PM, Dawany N, Dampier W, Byers SW, Pestell RG, Tozeren A. Bioinformatics analysis reveals transcriptome and microRNA signatures and drug repositioning targets for IBD and other autoimmune diseases. Inflamm Bowel Dis. 2012;18(12):2315–2333. doi: 10.1002/ibd.22958. [DOI] [PubMed] [Google Scholar]
- 106.Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H, Uchiyama K, Handa O, Kokura S, Ichikawa H, Yoshikawa T. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol. 2010;25(Suppl 1):S129–S133. doi: 10.1111/j.1440-1746.2009.06216.x. [DOI] [PubMed] [Google Scholar]
- 107.Bingham B, Ajit SK, Blake DR, Samad TA. The molecular basis of pain and its clinical implications in rheumatology. Nat Clin Pract Rheumatol. 2009;5(1):28–37. doi: 10.1038/ncprheum0972. [DOI] [PubMed] [Google Scholar]
- 108.Buchheit T, Van de Ven T, Shaw A. Epigenetics and the transition from acute to chronic pain. Pain Med. 2012 doi: 10.1111/j.1526-4637.2012.01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He Y, Wang ZJ. Let-7 microRNAs and Opioid Tolerance. Front Genet. 2012;3:110. doi: 10.3389/fgene.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Im YB, Jee MK, Jung JS, Choi JI, Jang JH, Kang SK. miR23b ameliorates neuropathic pain in spinal cord by silencing NADPH oxidase 4. Antioxid Redox Signal. 2012;16(10):1046–1060. doi: 10.1089/ars.2011.4224. [DOI] [PubMed] [Google Scholar]
- 111.Kynast KL, Russe OQ, Moser CV, Geisslinger G, Niederberger E. Modulation of central nervous system-specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain. 2012 doi: 10.1016/j.pain.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 112.Sengupta JN, Pochiraju S, Kannampalli P, Bruckert M, Addya S, Yadav P, Miranda A, Shaker R, Banerjee B. MicroRNA-mediated GABA(Aalpha-1) receptor subunit down-regulation in adult spinal cord following neonatal cystitis-induced chronic visceral pain in rats. Pain. 2013;154(1):59–70. doi: 10.1016/j.pain.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK. MicroRNA modulation in complex regional pain syndrome. J Transl Med. 2011;9:195. doi: 10.1186/1479-5876-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 115.Ding Z, Wang X, Khaidakov M, Liu S, Mehta JL. MicroRNA hsa-let-7 g targets lectin-like oxidized low-density lipoprotein receptor-1 expression and inhibits apoptosis in human smooth muscle cells. Exp Biol Med (Maywood) 2012;237(9):1093–1100. doi: 10.1258/ebm.2012.012082. [DOI] [PubMed] [Google Scholar]
- 116.Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX, Ye QH, Yu J, Shi X, Tang ZY, Wang XW. Let-7 g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52(5):690–697. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang K, Lin ZQ, Long B, Li JH, Zhou J, Li PF. Cardiac hypertrophy is positively regulated by MicroRNA miR-23a. J Biol Chem. 2012;287(1):589–599. doi: 10.1074/jbc.M111.266940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wada S, Kato Y, Okutsu M, Miyaki S, Suzuki K, Yan Z, Schiaffino S, Asahara H, Ushida T, Akimoto T. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem. 2011;286(44):38456–38465. doi: 10.1074/jbc.M111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L, Gemma A. MiR-23a regulates TGF-beta-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. 2012;41(3):869–875. doi: 10.3892/ijo.2012.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 121.Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50(2):575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 122.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W, Shi Y, Harley JB, Shen N, Qian Y. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB 2, TAB 3 and IKK-alpha. Nat Med. 2012;18(7):1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 123.Huang Z, Chen X, Yu B, He J, Chen D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem Biophys Res Commun. 2012;423(2):265–269. doi: 10.1016/j.bbrc.2012.05.106. [DOI] [PubMed] [Google Scholar]
- 124.Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep. 2012;27(2):594–598. doi: 10.3892/or.2011.1530. [DOI] [PubMed] [Google Scholar]
- 125.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55(2):465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS, Spin JM. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226(4):1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J Biol Chem. 2010;285(38):29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang Z, Huang S, Wang Q, Liang L, Ni S, Wang L, Sheng W, He X, Du X. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011;71(7):2582–2589. doi: 10.1158/0008-5472.CAN-10-3032. [DOI] [PubMed] [Google Scholar]
- 130.Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, Sudo S, Ju J, Sakuragi N. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2012 doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J Biol Chem. 2012;287(25):21093–21101. doi: 10.1074/jbc.M111.330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J, Ye Y, Wang Q, Long Z, Zhou Y, Du C, He X, Shi Y. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17(24):7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 133.Aprelikova O, Palla J, Hibler B, Yu X, Greer YE, Yi M, Stephens R, Maxwell GL, Jazaeri A, Risinger JI, Rubin JS, Niederhuber J. Silencing of miR-148a in cancer-associated fibroblasts results in WNT10B-mediated stimulation of tumor cell motility. Oncogene. 2012 doi: 10.1038/onc.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou L, Qi X, Potashkin JA, Abdul-Karim FW, Gorodeski GI. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem. 2008;283(42):28274–28286. doi: 10.1074/jbc.M802663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104(9):3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23(3):458–469. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Song XW, Li Q, Lin L, Wang XC, Li DF, Wang GK, Ren AJ, Wang YR, Qin YW, Yuan WJ, Jing Q. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol. 2010;225(2):437–443. doi: 10.1002/jcp.22217. [DOI] [PubMed] [Google Scholar]
- 138.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283(26):18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 139.Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, Yang AG, Zhang R. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun. 2012;420(4):787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 140.Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, Zhang Q, Dong L, Dong J. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep. 2012;27(3):685–694. doi: 10.3892/or.2011.1561. [DOI] [PubMed] [Google Scholar]
- 141.Sepramaniam S, Armugam A, Lim KY, Karolina DS, Swaminathan P, Tan JR, Jeyaseelan K. MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J Biol Chem. 2010;285(38):29223–29230. doi: 10.1074/jbc.M110.144576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Macconi D, Tomasoni S, Romagnani P, Trionfini P, Sangalli F, Mazzinghi B, Rizzo P, Lazzeri E, Abbate M, Remuzzi G, Benigni A. MicroRNA-324-3p Promotes Renal Fibrosis and Is a Target of ACE Inhibition. J Am Soc Nephrol. 2012;23(9):1496–1505. doi: 10.1681/ASN.2011121144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guo L, Qiu Z, Wei L, Yu X, Gao X, Jiang S, Tian H, Jiang C, Zhu D. The microRNA-328 regulates hypoxic pulmonary hypertension by targeting at insulin growth factor 1 receptor and L-type calcium channel-alpha1C. Hypertension. 2012;59(5):1006–1013. doi: 10.1161/HYPERTENSIONAHA.111.185413. [DOI] [PubMed] [Google Scholar]
- 144.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, Gupta M. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem. 2012;287(16):12913–12926. doi: 10.1074/jbc.M111.331751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, Yu Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23(20):2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ueno K, Hirata H, Majid S, Yamamura S, Shahryari V, Tabatabai ZL, Hinoda Y, Dahiya R. Tumor suppressor microRNA-493 decreases cell motility and migration ability in human bladder cancer cells by downregulating RhoC and FZD4. Mol Cancer Ther. 2012;11(1):244–253. doi: 10.1158/1535-7163.MCT-11-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Okamoto K, Ishiguro T, Midorikawa Y, Ohata H, Izumiya M, Tsuchiya N, Sato A, Sakai H, Nakagama H. miR-493 induction during carcinogenesis blocks metastatic settlement of colon cancer cells in liver. EMBO J. 2012;31(7):1752–1763. doi: 10.1038/emboj.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rieder F, Kessler SP, West GA, Bhilocha S, de la Motte C, Sadler TM, Gopalan B, Stylianou E, Fiocchi C. Inflammation-induced endothelial-to-mesenchymal transition: a novel mechanism of intestinal fibrosis. Am J Pathol. 2011;179(5):2660–2673. doi: 10.1016/j.ajpath.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Richter B, Roslind A, Hesse U, Nordling J, Johansen JS, Horn T, Hansen AB. YKL-40 and mast cells are associated with detrusor fibrosis in patients diagnosed with bladder pain syndrome/interstitial cystitis according to the 2008 criteria of the European Society for the Study of Interstitial Cystitis. Histopathology. 2010;57(3):371–383. doi: 10.1111/j.1365-2559.2010.03640.x. [DOI] [PubMed] [Google Scholar]
- 150.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH (2008) MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 135 (5):1624-1635 e1624. doi:10.1053/j.gastro.2008.07.068 [DOI] [PubMed]
- 151.Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16(10):1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sanchez-Simon FM, Zhang XX, Loh HH, Law PY, Rodriguez RE. Morphine regulates dopaminergic neuron differentiation via miR-133b. Mol Pharmacol. 2010;78(5):935–942. doi: 10.1124/mol.110.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, Brant SR, Kwon JH. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17(1):241–250. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci USA. 2011;108(20):8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, Li XP. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 156.Ma Y, Yu S, Zhao W, Lu Z, Chen J. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett. 2010;298(2):150–158. doi: 10.1016/j.canlet.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 157.Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, Soong R. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer. 2010;46(8):1456–1463. doi: 10.1016/j.ejca.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 158.Patron JP, Fendler A, Bild M, Jung U, Muller H, Arntzen MO, Piso C, Stephan C, Thiede B, Mollenkopf HJ, Jung K, Kaufmann SH, Schreiber J. MiR-133b targets antiapoptotic genes and enhances death receptor-induced apoptosis. PLoS ONE. 2012;7(4):e35345. doi: 10.1371/journal.pone.0035345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xie L, Ushmorov A, Leithauser F, Guan H, Steidl C, Farbinger J, Pelzer C, Vogel MJ, Maier HJ, Gascoyne RD, Moller P, Wirth T. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119(15):3503–3511. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 160.Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X, Tang H. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279(7):1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 161.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1 alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104(7):879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, Beckebaum S. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Haenisch S, Laechelt S, Bruckmueller H, Werk A, Noack A, Bruhn O, Remmler C, Cascorbi I. Down-regulation of ATP-binding cassette C2 protein expression in HepG2 cells after rifampicin treatment is mediated by microRNA-379. Mol Pharmacol. 2011;80(2):314–320. doi: 10.1124/mol.110.070714. [DOI] [PubMed] [Google Scholar]
- 164.Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I, Vyzula R, Slaby O. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16(11):2655–2666. doi: 10.1111/j.1582-4934.2012.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]