Abstract
This review examines the role of drug metabolism and drug target polymorphism in determining the clinical response to antidepressants. Even though antidepressants are the most effective available treatment for depressive disorders, there is still substantial need for improvement due to the slow onset of appreciable clinical improvement and the association with side effects. Moreover, a substantial group of patients receiving antidepressant therapy does not achieve remission or fails to respond entirely. Even if the large variation in antidepressant treatment outcome across individuals remains poorly understood, one possible source of this variation in treatment outcome are genetic differences. The review focuses on a few polymorphisms which have been extensively studied, while reporting a more comprehensive reference to the existing literature in table format. It is relatively easy to predict the effect of polymorphisms in drug metabolizing enzymes, such as cytochromes P450 2D6 (CYP2D6) and cytochrome P450 2C19 (CYP2C19), which may be determined in the clinical context in order to explain or prevent serious adverse effects. The role of target polymorphism, however, is much more difficult to establish and may be more relevant for disease susceptibility and presentation rather than for response to therapy.
Keywords: Polymorphisms, Pharmacokinetics, Pharmacodynamics, Antidepressant treatment, Adverse effects, Suicidal behavior
Introduction
Depression is a complex psychiatric disorder affecting approximately 15 % of the population with high morbidity and mortality. Despite the high prevalence and socioeconomic impact of depressive illnesses, little is known about their etiology. The underlying causes of most mood disorders remain asyet unknown. Heritability estimates based on family aggregation and contrasting monozygotic and dizygotic twin studies indicate up to a 50 % genetic component [1]. This strong inheritable component of psychiatric illnesses when coupled with environmental influences results in increased susceptibility to develop the illness in response to stressful and/or adverse enviromental conditions.
In the 1960s, the observation that depressive symptoms are influenced by pharmacological manipulation of monoamines led to the hypothesis that depression results from reduced availability or functional deficiency of monoaminergic transmitters in certain cerebral regions. There are, however, limitations to current monoamine theories related to mood disorders. In the last few years, a growing body of experimental evidence has shown that other classes of endogenous compounds, such as neuropeptides and amino acids, are likely to play a significant role in the pathophysiology of affective disorders. Furthermore, as neuroscience has developed, neuronal networks and intracellular pathways have been identified and characterized, describing the interaction between monoamines and receptors able to modulate the expression of intracellular proteins and neurotrophic factors (for a review, see [2]).
Even though antidepressants are the most effective treatment available for depressive disorders, there is still substantial need for improvement due to the slow onset of appreciable clinical improvement and associated side effects. Moreover, a substantial group of patients receiving antidepressant therapy does not achieve remission or fails to respond entirely. Even if the large variation in antidepressant treatment outcome across individuals remains poorly understood, one possible source of this variations are genetic differences [3]. Pharmacogenetics investigates the influence of genes on the pharmacokinetics and pharmacodynamics of drugs [4]. In recent years, a number of pharmacogenetic studies on antidepressant drugs have been published and several polymorphisms in candidate genes have been associated with antidepressant response [5, 6]. However, few pharmacogenetic associations have been replicated, and, those that have, explain only a small fraction of individual differences in antidepressant response.
Pharmacokinetic aspects
The term “pharmacokinetics” refers to processes such as absorption, distribution, metabolism, and elimination (ADME) that influence the delivery of a drug to its target(s). Several genetic polymorphisms in key regulatory genes involved in drug metabolism, such as the cytochrome P450 (CYP) gene family, and drug transporter molecules like genes from the multidrug resistance (MDR) gene family, have been reported to influence the pharmacokinetics of drugs [7]. These gene families are also implicated in the pharmacokinetics of antidepressant drugs [8]. We will focus on the cytochromes P450 CYP2D6 and CYP2C19, and on p-glycoprotein, one member of the MDR gene family, as they have been most extensively investigated.
Polymorphisms linked to the ADME system
CYP2D6
Most antidepressants are metabolized by CYP2D6, which accounts for a small percentage of all hepatic CYP enzymes, but is responsible for the metabolism of approximately 25 % of all drugs metabolized by CYP [9]. The CYP2D6 gene is highly polymorphic with more than 60 allele variants [http://www.cypalleles.ki.se/cyp2d6.htm]. CYP2D6*4 is the most common allele variant (allele frequency of 20 %) in Caucasians and is the most frequent nonfunctional allele in the poor metabolizer (PM) phenotype; over 75 % of PMs are carriers of this polymorphism [10]. Genetic variations of the CYP2D6 gene have been associated with predictable variations in serum levels of antidepressant medication [11] and clinical responses [12, 13].
Several studies have investigated the relationship between plasma levels of antidepressant drugs and CYP2D6 polymorphisms, and case reports have suggested an association between such variants and antidepressant-related side effects [14, 15]. Knowledge of CYP2D6 metabolizer status could be helpful in personalizing dose escalation schemes for certain antidepressants, since most tricyclic antidepressants (TCAs) are substrates of CYP2D6, and a series of newer antidepressant drugs, especially the selective serotonin reuptake inhibitors (SSRIs) fluoxetine, fluvoxamine, and paroxetine, are strong inhibitors of its activity. In particular, due to the absence of CYP2D6-mediated metabolism, poor metabolizers (PMs) have higher plasma concentrations of antidepressants metabolized by CYP2D6 than extensive metabolizers (EMs), and are, therefore, more likely to suffer from dose-dependent adverse drug reactions (ADRs). PMs receiving TCA treatment may be particularly at risk for cardiotoxicity and other severe ADRs, because TCAs have a narrow therapeutic range. Severe ADRs require dose reductions or discontinuation of antidepressant therapy. In patients receiving a fixed dose of venlafaxine, a selective serotonin–norepinephrine reuptake inhibitor (SNRI), with a dosage of 75 mg/day, those who lacked a fully active CYP2D6 allele were not able to tolerate a maintenance dosage higher than 75 mg/day when compared to patients with at least one fully active 2D6 allele [16]. This finding was further supported by data demonstrating that, in users of TCA, the chance of switching to any other antidepressant within 45 days is significantly higher in PMs than in EMs [17, 18]. In contrast, the CYP2D6 genotype had no effect on paroxetine and mirtazapine discontinuations and adverse events in an 8-week long, double-blind randomized study on antidepressant intolerance [19]. Therefore, while an identification of PM may prevent overdosing and the occurrence of specific side effects with TCAs and MAO-Is and possibly cardiotoxic side effects of venlafaxine, this seems to be less relevant for other antidepressant drug classes, including SSRIs.
CYP2C19
CYP2C19 is a polymorphic enzyme active in the metabolism of clinically used drugs, several of which are psychoactive such as the antidepressants sertraline [20], escitalopram [21] and citalopram [22], as well as diazepam [23]. CYP2C19*2 and *17 are the most studied variant alleles. In Europeans and Asians, the defective CYP2C19*2 allele is responsible for the majority of the PM phenotypes [115]. The CYP2C19*17 allele is associated with a rapid metabolizer (RM) phenotype in vivo for various drugs, including the antidepressant escitalopram, and has an allele frequency of 18–27 % in different European populations [21, 24]. In particular, data from Rudberg et al. [21] demonstrated a strong gene dose effect on the steady state plasma level of escitalopram in relation to the CYP2C19 genotype in a psychiatric population, with a 15-fold difference in escitalopram concentration between subjects homozygous for CYP2C19*17 (RMs) and subjects homozygous for defective CYP2C19 alleles (PMs). Interestingly enough, Sim et al. [24] recently demonstrated that subjects of European ancestry with a CYP2C19*2 PM genotype have a significantly lower level of depressive symptoms than EMs. The mechanism underlying such association has yet to be elucidated. A possible link between CYP2C19 and serotonin (5-HT) has only been described in vitro, where CYP2C19 has been suggested to participate in the biotransformation of 5-HT [25]. However, further studies are needed in order to investigate the functional link between CYP2C19 and depressive symptoms. It would be particularly important to understand whether the association is due to CYP2C19’s activity in the periphery or in the brain toward endogenous neurotransmitters and neuromodulators.
There is still debate whether therapeutic efficacy may be improved and/or adverse effects could be prevented by the use of genotyping procedures, particularly considering that genotypes often do not correspond to a well-defined phenotype. However, the approval by the FDA of the pharmacogenetic test, the AmpliCyp CYP450 Test (Roche Molecular System), which asseses both polymorphic genes CYP2D6 and CYP2C19, may be of help to validate studies on personalized therapy of depression.
P-glycoprotein
P-glycoprotein is a member of the highly conserved superfamily of ATP-binding cassette (ABC) transporter proteins [26]. P-glycoprotein is a 170-kDa glycoprotein encoded by the ABCB1 gene (alternate name MDR1) on chromosome 7q21 [27] which protects cells throughout the healthy organism against many drugs by acting as an efflux pump for xenobiotics. Additionally, the location of P-glycoprotein at the blood–brain barrier (BBB) assigns it an important role in regulating the concentration of psychotropic and antidepressant drugs in the brain [28, 29]. In the case of paroxetine, venlafaxine, and fluoxetinen the data indicate that they might be ligands of P-glycoprotein, while in the case of citalopramn the data are conflicting [30, 31]. Hence, functional polymorphisms in this gene may influence intracerebral concentration of specific antidepressants, by altering the efficiency with which P-glycoprotein transports substrate antidepressants at the BBB. Within this context, the literature reports the association of three polymorphisms, two synonymous SNPs (C3435T -rs1045642- and C1236 -rs1128503-) and one non-synonymous SNP (G2677T -rs2032582-), with altered P-glycoprotein activity, which is responsible for differences in drug (venlafaxine and paroxetine) plasma levels of ligands [32]. The G2677T SNP has been reported to alter activity and ligand specificity of P-glycoprotein [33], even though this finding has not been further replicated [34]. Indeed, functional significance has also been attributed to the two synonymous SNPs, C3435T and C1236 [34]. In general, studies that have investigated the influence of these functional polymorphisms on antidepressant drug plasma levels, side effects profile, and clinical response have reported contradictory findings to date [35–37]. Further replication and prospective studies will be important to determine whether the knowledge of ABCB1 status may be of clinical relevance in the administration of an antidepressant.
Pharmacodynamic aspects
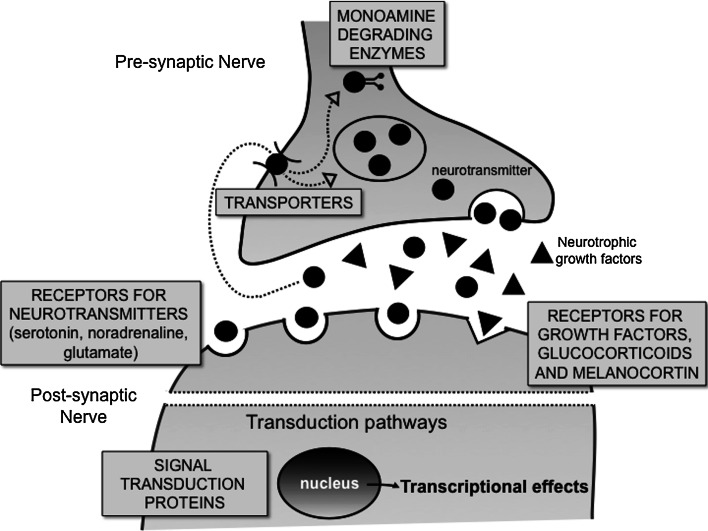
The genetics of pharmacodynamics relates to genes directly involved in antidepressant drug effects. In contrast to the pharmacokinetics of antidepressants, for which at least the major mechanisms have been uncovered, the pharmacodynamics of antidepressants is still unclear. The major current hypotheses for antidepressant activity are normalization of a relative monoamine deficiency, of stress hormone system hyperactivity, or of decreased neurotrophic activity [2, 38]. Multiple targets have been linked to depression (for a detailed overview, see Table 1). As detailed below, we will focus on the serotonin transporter gene SLC6A4 and on tryptophan hydroxylase, as they have been most extensively investigated. The other targets, their clinical relevance, and the antidepressants involved are summarized in Table 1. The table reports all the target polymorphisms which are described as clinically relevant and reach statistical significance in the published literature, as well as the antidepressants involved. The most relevant are discussed in the following text. The table legend reports a classification of the involved antidepressants that may help the reader to follow the table data and the discussion. The table is organized according to the target or target class involved (summarized in Fig. 1).
Table 1.
Target polymorphims published as clinically relevant and antidepressants involved
| Target class | Polymorphisms investigated | Region | Published as clinically relevant and antidepressant involved | References |
|---|---|---|---|---|
| Transporters | ||||
| SLC6A4 (serotonin transporter) | rs25531 | Promoter | Patients with MDD carrying the rs25531(A) allele may be at increased risk for side effects (e.g. headaches), when treated with escitalopram 10–20 mg/day for 12 weeks | [39] |
| 44 bp Del/Ins | Promoter | Significant association of the l variant of 5-HTTLPR with a better response to SSRIs (fluvoxamine, paroxetine, fluoxetine, and citalopram). This effect seemed independent from ethnic differences. The subjects with the s/s genotype have difficulties to reach remission, and take a long time, over 4 weeks, to respond, as do the subjects with the S allele | [40] | |
|
S allele carriers treated with paroxetine (20–40 mg/day) for 8 weeks experienced more severe adverse events. Among mirtazapine (15–45 mg/day)-treated patients, S allele carriers had fewer discontinuations due to adverse events |
[41] | |||
| Association between the l carriers and reduced side-effect rate under SSRI treatment (fluvoxamine, paroxetine, fluoxetine, and citalopram) | [5] | |||
| SLC6A2 (norepinephrine transporter) | rs5569 | Coding exon | In Japanese patients with MDD treated with milnacipran (50–100 mg/day, for 6 weeks), a dual serotonin/norepinephrine reuptake inhibitor, the T allele was associated with a superior antidepressant response, whereas the AA genotype was related to a slower onset of therapeutic response | [42] |
| In a Korean sample, the GG genotype was associated with a better response to the NRI nortriptyline than to SSRIs (fluoxetine or sertraline) | [43] | |||
| rs2242446 | 5′ UTR | In Japanese subjects, the T allele was associated with a superior response to milnacipran (50–100 mg/day, for 6 weeks) | [44] | |
| No correlation between the T allele and SSRIs (fluoxetine, paroxetine, or citalopram) or venlafaxine response in an Asiatic population of depressed patients after 6 weeks of treatment | [45] | |||
| rs5564 | Intron | The G allele was associated with a better remission after 8 weeks antidepressant treatment with desipramine | [46] | |
| rs1362621 | Promoter | The A allele was associated with a better remission after 8 weeks antidepressant treatment with fluoxetine | [46] | |
| Monoamine degrading enzymes | ||||
| MAO-A | uVNTR | Promoter |
Patients carrying the short-form polymorphism had greater improvement, i.e., greater the reduction in HRSD score from baseline, higher is the remission rate, and lower the drop-out rate than patients with the long-form polymorphism after taking mirtazapine (30 mg/day) for 7 weeks. Furthermore, the MAOA 4R allele was associated with enhanced vulnerability to suicide in depressed males, but not in community subjects. |
[47, 48] |
| Receptors for neurotransmitters | ||||
| Serotonin | ||||
| 5-HT1A | rs6295 | Promoter | The CC genotype was positively associated with response to antidepressants fluoxetine or nefadozone combined with the 5-HT1A agonist flibanserin | [49] |
| rs10042486 | Promoter | The C allele was strongly associated with SSRI/SNRI response in a Japanese sample with major depression followed for 6 weeks | [50] | |
| rs1364043 | Downstream | The T allele was strongly associated with SSRI/SNRI response in Japanese patients with major depression followed for 6 weeks | [50] | |
| 5-HT2A | rs6311 | Promoter |
In a Korean population, the GG genotype appeared to be associated with a better response to citalopram, with especially the G allele being related to core depressive symptoms and psychic anxiety improvement. In Japanese patients affected by MDD, the GG genotype was associated with a good response to SSRIs, mainly to fluvoxamine, and significantly with severe nausea in paroxetine-treated patients. |
[51, 52] |
| rs6313 | Coding exon | Different treatment response in patients with one or two C alleles, with a significantly higher decrease in HAMD-17 after 4 weeks of antidepressant treatment (19 % TCAs, 17 % mirtazapine, 14 % SSRIs, 13 % electroconvulsive therapy, 13 % repetitive transcranial magnetic stimulation, 24 % combinations) | [53] | |
| 5-HT3A | rs1062613 | 5′ UTR | In Japanese patients affected by MDD, the CC genotype was associated with an antidepressant response to SSRIs, and more significantly in paroxetine-treated patients after 6 weeks treatment | [52] |
| 5-HT6 | rs1805054 | Coding exon | The CT genotype had significantly better treatment response (venlafaxine and fluoxetine for 8 weeks) than the homozygote group (CC + TT genotypes), mainly in the somatic-anxiety subcategory and the total score of HAM-D. This finding has not been replicated by further studies | [54, 55] |
| Noradrenaline | ||||
| ADRA2A (adrenoreceptor alpha-2) | rs11195419 | 3′UTR | Among men taking nortriptyline, enrolled in the Genome-based Therapeutic Drugs for Depression (GENDEP) study, the A allele was associated with new-onset or worsening suicidal ideation | [56] |
| rs1800544 | Promoter | The C allele carriers showed a better improvement than GG subjects treatment with milnacipran | [57] | |
| Glutamate | ||||
| GRIA3 (glutamate receptor, ionotrophic, AMPA 3) | rs4825476 | Intron | The G allele has been linked to increased thoughts of suicide in patients taking the antidepressant drug citalopram. The increased risk was calculated to be 1.9× | [58] |
| GRIK2 (glutamate receptor, ionotropic kainate 2) | rs2518224 | Intron | The C allele has been linked to increased thoughts of suicide in patients using the antidepressant drug citalopram. The risk is only seen for individuals CC homozygotes. The odds ratio for these individuals was 8.2 | [58] |
| GRIK4 (glutamate receptor, ionotropic, kainate 4) | rs1954787 | Intron | The C allele was associated with an increased rate of successful response to treatment of depression with the drug citalopram | [59, 60] |
| Receptors for glucocorticoids, melanocortin, and corticotropin-releasing hormone | ||||
| GR (glucocorticoid receptor) | ER22/23EK | Transactivation domain | Association between the ER22/23EK genotype and faster response to antidepressant (SSRIs, TCAs, or mirtazapine) after 2 weeks of treatment. Carriers of the ER22/23EK polymorphism remitted on average 5 days faster to antidepressant treatment than non-carriers | [61] |
| MC1R (melanocortin 1 receptor) | rs2228478 | No data | In Mexican-American patients with MDD, the A allele has been associated with the remission with desipramine treatment. No associations were found for remission with fluoxetine treatment or for the combined sample treated with fluoxetine or desipramine | [62] |
| rs2228479 | No data | In Mexican-American patients with MDD, the A allele has been associated with the remission with desipramine treatment. No associations were found for remission with fluoxetine treatment or for the combined sample treated with fluoxetine or desipramine | [62] | |
| CRHR1 (corticotropin releasing hormone receptor 1) | rs242941 | Intron |
In Han Chinese patients, the homozygous GAG haplotype was associated with fluoxetine (20 mg/day, for 6 weeks) therapeutic response in MDD patients of high-anxiety In Mexican-Americans, homozygosity for the GAG haplotype was associated with a 70 % greater reduction in HAM-A scores and 31 % greater reduction in HAM-D scores after treatment with fluoxetine (10–40 mg/day) or desipramine (50–200 mg/day) for 8 weeks |
[63, 64] |
| rs1876828 | Intron |
In Han Chinese patients, the homozygous GAG haplotype was associated with fluoxetine (20 mg/day, for 6 weeks) therapeutic response in MDD patients of high-anxiety. In Mexican-Americans, homozygosity for the GAG haplotype was associated with a 70 % greater reduction in HAM-A scores and 31 % greater reduction in HAM-D scores after treatment with fluoxetine (10–40 mg/day) or desipramine (50–200 mg/day) for 8 weeks |
[63, 64] | |
| rs242939 | Intron |
In Han Chinese patients, the GG genotype and homozygous GAG haplotype were associated with fluoxetine (20 mg/day, for 6 weeks) therapeutic response in MDD patients of high-anxiety In Mexican-Americans, homozygosity for the GAG haplotype was associated with a 70 % greater reduction in HAM-A scores and 31 % greater reduction in HAM-D scores after treatment with fluoxetine (10–40 mg/day) or desipramine (50–200 mg/day) for 8 weeks |
[63, 64] | |
| CRHR2 (corticotropin releasing hormone receptor 2) | rs2270007 | Intron | In Spanish depressed patients, G allele carriers showed a worse overall response to citalopram (20–40 mg/day, for 12 weeks) | [65] |
| Receptors for neurotrophic factors | ||||
| NTRK2 (neurotrophic tyrosine kinase, receptor, type 2) | rs2289657 | Coding exon | In Mexican-American patients with MDD, the C allele has been found to be associated with a better response to desipramine (50–200 mg/day, for 8 weeks) | [46] |
| rs56142442 | Coding exon | In Mexican-American patients with MDD, the C allele has been found to be associated to a better response to desipramine (50–200 mg/day, for 8 weeks) | [46] | |
| rs2289658 | Intron | In Mexican-American patients with MDD, treated with fluoxetine (10–40 mg/day), or desipramine (50–200 mg/day) for 8 weeks, the T allele has been found to be associated with a better antidepressant response | [46] | |
| rs45596934 | Intron | In Mexican-American patients with MDD, treated with fluoxetine (10–40 mg/day), or desipramine (50–200 mg/day) for 8 weeks, the A allele has been found to be associated with a better antidepressant response | [46] | |
| rs1624327 | Intron | In Mexican-American patients with MDD, treated with fluoxetine (10–40 mg/day), or desipramine (50–200 mg/day) for 8 weeks, the A allele has been found to be associated with a better antidepressant response | [46] | |
| Signal transduction proteins | ||||
| GNB3 (G protein beta 3 subunit) | rs5443 | Coding exon |
In Taiwanese patients with MDD, the C allele was associated with short-term antidepressant (venlafaxine and SSRIs including sertraline, paroxetine, and fluoxetine response, for 2 weeks). Carriers of the C allele responded better to treatment than those with the TT variant. In white European patients with moderate to severe major depression, the TT genotype moderated change in neurovegetative symptoms during nortriptyline treatment, but had no effect on the mood or cognitive symptoms of depression; this effect was marked in patients treated with the TCA nortriptyline but not significant in patients treated with the SSRI escitalopram |
[66, 67] |
| FKBP5 (FK506 binding protein 5) | rs1360780 | Intron | In Caucasian non-Hispanics, the T allele was associated with increased risk for depression, with an odds ratio of 1.39. Subjects with TT homozgyotes tend to report more depressive episodes, but they also respond better to treatment with antidepressants, including citalopram | [68, 69] |
| Growth factors | ||||
| BDNF (brain derived neurotrophic factor) | rs6265 | Coding exon |
In Chinese patients with MDD, treated with the SSRI fluoxetine (20 mg/day, for 6 weeks), the A allele was associated with lower fluoxetine side effects In a Caucasian sample with MDD, treated with different antidepressants (mirtazapine, citalopram/escitalopram, venlafaxine, TCAs and MAO-I), the G allele predicted worse treatment response over 6 weeks in clinical subtypes of depression such as melancholic depression only |
[70–72] |
| rs7103411 | Intron | In a Caucasian sample with MDD, treated with different antidepressants (mirtazapine, citalopram/escitalopram, venlafaxine, TCAs and MAO-I), the T allele predicted worse treatment response over 6 weeks in clinical subtypes of depression such as melancholic depression only | [71] | |
| rs10501087 | The C allele was associated with non-response to antidepressant treatment | [72] | ||
| rs7124442 | 3′UTR | In a Caucasian sample with MDD, treated with different antidepressants (mirtazapine, citalopram/escitalopram, venlafaxine, TCAs and MAO-I), the T allele has been linked to a worse treatment response over 6 weeks in clinical subtypes of depression | [71] | |
| rs61888800 | 5′ UTR | In Mexican-Americans with MDD, treated with fluoxetine (10–40 mg/day), or desipramine (50–200 mg/day) for 8 weeks, this SNP was found to be associated with the better response to antidepressant treatment. Patients who had the GG genotype showed a larger average reduction of HAM-D21 score of 66.3 % compared with those who had the non-GG genotype | [73] | |
The main antidepressant drugs are classified as: monoamine oxidase inhibitors (MAO-Is): phenelzine; reversible inhibitors of MAO-A (RIMA): moclobemide, toloxatone, pirlindole; tricyclic antidepressants (TCAs): imipramine, amitriptyline, clomipramine, desipramine, nortriptyline; selective serotonin reuptake inhibitors (SSRIs): citalopram, escitalopram, fluoxetine, fluvoxamine, paroxetine, sertraline; serotonin-norepinephrine reuptake inhibitors (SNRIs): venlafaxine, milnacipran; norepinephrine reuptake inhibitors (NRIs): atomoxetine, reboxetine, viloxazine; noradrenergic and specific serotonergic antidepressants (NaSSAs): mianserin, mirtazapine, setiptiline; serotonin antagonist and reuptake inhibitors (SARIs): trazodone, etoperidone, nefazodone; dopaminergic drugs: bupropion, amisulpride
Fig. 1.
Classes of targets associated with polymorphisms putatively influencing antidepressant response. The figure summarizes the classes of targets whose polymorphisms have been related to antidepressant response. The literature and the findings are detailed in Table 1 and in the text
Polymorphisms of target
Serotonin transporter gene SLC6A4
The serotonin transporter (SLC6A4) regulates brain serotonin neurotransmission by transporting the neurotransmitter serotonin from synaptic space to presynaptic terminals. SLC6A4 is the principal site of action of many antidepressants (mainly SSRI, SNRI, and TCA) and it is a target of primary interest in the pharmacogenetics of depression. The gene encoding for the serotonin transporter (chromosome 17q11–12) shows a functional polymorphism in the promoter region (5-HTTLPR). The 5-HTTLPR is polymorphic because of the insertion/deletion of units 6–8, which produces a short (s) allele that is 44 bp shorter than the long (l) allele. In the baseline state, the long allele has twice the expression of the short variant, the latter being associated with low transcriptional efficiency, leading to decreased serotonin transporter expression [74]. The reduced efficiency of the SLC6A4 gene has been hypothesized as a risk factor that may explain some of the differences in clinical response to drugs acting at this receptor. Several studies have investigated the effect of this polymorphism on response to antidepressant treatment, with most studies focusing on SSRI treatment. Serretti et al. [40] showed a better response between the s/s variant of 5-HTTLPR and the remission rate under SSRI treatment (fluvoxamine, paroxetine, fluoxetine, and citalopram) and between both s/s and s/l variants and the response rate, even though a more recent meta-analysis performed by Taylor et al. [75] failed to replicate this result. Other studies have evaluated the association of 5-HTTLPR and remission in response to citalopram in the STAR*D sample [76]. The studies report different results, with some of them showing no association of 5-HTTLPR with remission under antidepressant treatment in the whole sample [77], while Mrazek et al. [76] replicated the association of the l-allele with better treatment outcome only in white non-Hispanic patients in the STAR*D sample. Changes in therapeutic response may be associated with changes in the impact of side effects. The meta-analysis by Kato and Serretti [5] concluded a reduced side-effect rate for the l-carriers (l/l and l/s). The putative predictive role of 5HTTLPR on side effects is thought to be limited to SSRIs, as in a study with mirtazapine, the l-carriers revealed more side effects under this drug [41]. More recently, rs25531, a biallelic polymorphism located in the sixth repeat of 5-HTTLPR, has been demonstrated to impact the response to antidepressant treatment, thus modulating the effect of the other 5-HTTLPR alleles [78]. The results, however, are still controversial with both negative [39, 79] and positive findings [76, 80]. More recent reports (as the GENDEP study [81]) were not able to confirm the modulatory role of rs25531 on SSRI response. Additional studies are required to obtain validated conclusions. On the other hand, on a theoretical basis, one would not expect that changes in the expression of the transporter would dramatically affect the clinical response unless the affinity for the inhibitors is changed, a feature which may be associated with changes in the sequence rather than in the expression of the transporter protein. At clinically active concentrations, antidepressants should be able to inhibit the transporters even if over-expressed. Changes in transporter expression may rather be associated with changes in the susceptibility to develop depression [82, 83].
Tryptophan hydroxylase
Serotonin is synthesized from tryptophan through the enzyme tryptophan hydroxylase (TPH) which occurs in two isoforms, TPH1 and TPH2. TPH1 is expressed in the central nervous system (CNS) as well as in tissues that express serotonin, such as skin, gut, and pineal gland. TPH2 is exclusively expressed in neuronal cell types and is the predominant isoform in the CNS. The TPH1 gene carries a DNA-variant 218A/C (rs1800532) in a potential transcription-factor binding site with a putative effect on gene expression. Several studies found an association between TPH1 polymorphisms and suicidal behavior and worse response to SSRIs [84], even though these data were not replicated in studies that followed [55, 85]. Some studies found an association between the 218A/C polymorphism and antidepressant treatment, with the CC genotype shown to predict beneficial response [5]. Associations between rs1800532 and side-effect profiles have also been found, with the AA genotype of TPH1 rs18532 significantly associated with bodyweight gain during antidepressive treatment [86].
THP2 has been implicated in the pathogenesis of major depressive disorders and in the mechanism of antidepressant actions. TPH2 genetic variants have been reported to play a role in acute therapeutic response to SSRIs. The variants rs1843809, rs1386494, and rs1487276 were found to be positively associated with response to fluoxetine [87], whereas none of these variants was correlated to citalopram response [88]. The inability of the polymorphism to equally affect the response to all representatives of the same class of antidepressants may reflect either different clinical experimental settings, small numbers of patients, or the fact that the action of the antidepressant relies on several contributing mechanisms which may differ for the various agents, in spite of the them sharing a common target. Although these results need to be clearly replicated, overall they suggest a role for the TPH2 gene in the pharmacogenetics of depression.
Pharmacogenetics of adverse effects and suicidal behavior upon antidepressant treatment
For nearly two decades, there has been concern that SSRIs and other antidepressants may induce an increased risk for suicidal ideation and behavior in some depressed patients [89–91]. The ability to identify which depressed individuals exposed to antidepressants are most likely to become suicidal would be very useful in the clinical setting. Pharmacogenomics may improve the identification of patients most likely to experience a suicidal attempt and may define clinical phenotypes associated with a lack of response and treatment-emergent suicidal events [92]. Associations between suicide events and genetic variations in the cAMP response element binding protein 1 gene (CREB1), brain-derived neurotrophic factor (BDNF), neurotrophic tyrosine kinase receptor type 2 (NTRK2), glutamatergic receptors, and FK506 binding protein 5 (FKBP5) are most consistent with a general increased genetic predisposition to suicidal behavior.
Polymorphisms in the CREB1 gene have been shown to predict suicidal events in depressed males treated with citalopram. This gene is involved in antidepressant responses [93], and its expression and phosphorylation are altered in post-mortem studies of suicide victims [94, 95]. However, the association between CREB1 and past suicide attempts has not recently been confirmed [96].
Brain-derived neurotrophic factor is thought to be involved in the etiology of depression as it plays a role in survival of neurons during hippocampal development. Altered expression of BDNF has been shown in post-mortem studies of suicides [97]. Antidepressant drugs increased expression of proteins associated with synaptic plasticity [98]. A single nucleotide polymorphism (SNP) was identified in the coding exon of the BDNF gene resulting in a valine (Val) to methionine (Met) change at amino acid position 66 (Val66Met) [99]. This polymorphism has been demonstrated to be correlated with decreased hippocampal volume and abnormal hippocampal activation, which may underlie an enhanced vulnerability [100]. The Val66meMet polymorphism has been shown to be related to suicide attempts in depressed and bipolar patients and in subjects with lower serotonin transporter availability [101, 102]. Also, a receptor for BDNF, NTRK2, has been linked to suicidal behavior and is also supported by post-mortem and association studies [97, 103].
FKBP5 is located on chromosome 6p21, a chromosomal region associated with bipolar disorder and psychosis [104], and is mainly expressed in the brain and in a wide range of human cell tissue, including muscle, liver, and thymus. The heatshock protein FKBP5 is a part of the mature GR heterocomplex reducing the receptor affinity for cortisol and is thought to diminish the binding of dynein to the complex and its nuclear translocation [105]. Interest in this protein has recently been reinforced by the observation that genetic variants of FKBP5 have been reported to be associated with adaptive changes in HPA-regulation and to be related to a different antidepressant response, pointing to FKBP5 as an important target for further studies of depression and treatment response [106]. A strong association between the complementary FKBP5 alleles and the risk for suicide attempts has recently been found in African-Americans [107].
A link between polymorphisms in glutamatergic receptors and suicidal events has also been reported. Both the GRIK2 and GRIA3 genes encode for ionotropic glutamate receptors, which have been implicated in the etiology of depression [108], suicide, and suicidal behavior [109]. Ketamine, an agonist for the ionotropic glutamate receptor NMDA, has been reported to reduce suicidal ideation and depressive symptoms in treatment-resistant patients [110], an observation which suggests that genetic association may indicate new ways of intervention to better tailor drug treatment and may provide suggestions for new drug association therapy in depression.
Conclusions
One of the essential objectives of modern pharmacotherapy of psychiatric patients should be to help the substantial proportion of patients who do not respond sufficiently or suffer adverse effects during treatment. Variable response and adverse effects may be explained by genetic variability in relation to pharmacodynamic and pharmacokinetic pathways, as well as by many other non-genetic factors, including misclassification of disease or subtype, cultural or environmental factors. Some of these variables may be amenable to control. Among them, pharmacogenetics holds a great promise for future clinical practice through the use of genetic markers for prediction of treatment outcome.
However, by analyzing data from the literature, several points have to be discussed. Sample sizes of pharmacogenetic studies are often very small so as to reach a validated result. Considerable heterogeneity is present among individual studies when considering population characteristics, type of intervention, outcome measurement, and robustness of the results. Ethnicity of the patient sample might play a role in determining the direction of the effect. As an example regarding the serotonin transporter, in Caucasian patients response to SSRIs seemed less favorable for patients with the s/s genotype than for those with the s/l and l/l genotypes [111]. Moreover, differences in diagnosis could affect interpatient comparability. In particular, different versions of the Hamilton Rating Scale for Depression (HAM-D) have been used in the individual studies analyzed, which suggests that the version used or the experience of the interviewers using the HAM-D may influence depression scores. Different classes of antidepressants and different agents of the same class were used in the analyzed studies, with some studies using multiple dosages of drugs and others that did not specify the dosage used. Hence, within this context, using different antidepressants and varying dosages, a comparison of study results is precluded.
A SWOT (strengths, weaknesses, opportunities, threats) analysis of the pharmacogenetic studies applied to antidepressant therapy is reported in Table 2. On these grounds, it is difficult to ascertain the present clinical value of pharmacogenetics in the pharmacotherapy of depression. It is tempting to speculate that the usual clinical practice and attention to the evolution of symptoms in the patient may efficiently subsume a more sophisticated approach. On the other hand, pharmacogenetic studies have been useful in post hoc analysis to unravel cases in which a standard dose of some antidepressants, mostly tricyclics, may have led to severe or catastrophic side effects in poor metabolizers, or to complete unresponsivness in extensive metabolizers [112]. Moreover, target polymorphisms have led to the discovery of new targets or to targets for associated therapies aimed to solve therapy-associated problems, such as suicidal behavior, as in the case of ketamine trials [110, 113]. Even if all the observations mentioned above apply to clinical research rather than to present clinical practice, they may lead to the development of new molecular diagnostic tools with a favorable cost-effectiveness profile. It is easy to foresee that such tools should combine several relevant polymorphisms in the same assay at an affordable cost. It is also important to develop methods to understand how complex phenotypes arise from individual polymorphisms and their interactions, a challenge that computational biology approaches have already begun to face in simple organisms [114].
Table 2.
SWOT analysis of the pharmacogenetic studies applied to antidepressant therapy
| Strengths | Weaknesses |
|
To help the proportion of patients not responding sufficiently or suffering adverse effects during treatment To use genetic markers for prediction of treatment outcome |
Inadeguate sample size Lack of replication Differences in diagnosis (linked to different versions of the HAM-D used) Use of different classes of antidepressants and/or different agents of same class Differences in dosages |
| Opportunities | Threats |
|
Discovery of clinically useful predictors Shedding light on mechanisms of drug action Identification of new targets not directly involved in mechanism of action |
Phenotype definition Diagnostic uncertainty |
Acknowledgments
The review was written as partially based on the work of the students of 2011 IUSS (Istituto Universitario di Studi Superiori) Course on Pharmacogenetics of the University of Pavia. The partecipating students, who collected various materials on the pharmacogenetics of psychiatric drugs, were: Aitala Becherucci Edoardo, Aloisio Elena, Azzolini Maria, Barone Gisella, Chiodaroli Elena, Conte Marco, Danesi Daniela, Di Lodovico Laura, Gavuzzi Marta, Giorgio Alessandro, Gitto Salvatore, Melotti Dario, Pelizzari Giacomo, Peluso Francesca, Pepe Antonella, Russo Giulia, Schiavi Susanna, and Vegezzi Elisa.
References
- 1.Hamet P, Tremblay J. Genetics and genomics of depression. Metabolism. 2005;54:10–15. doi: 10.1016/j.metabol.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Lanni C, Govoni S, Lucchelli A, Boselli C. Depression and antidepressants: molecular and cellular aspects. Cell Mol Life Sci. 2009;66:2985–3008. doi: 10.1007/s00018-009-0055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franchini L, Serretti A, Gasperini M, Smeraldi E. Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees. J Psychiatr Res. 1998;32:255–259. doi: 10.1016/S0022-3956(98)00004-1. [DOI] [PubMed] [Google Scholar]
- 4.Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry. 2010;15:473–500. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- 6.Uher R, Huezo-Diaz P, Perroud N, Smith R, Rietschel M, Mors O, Hauser J, Maier W, Kozel D, Henigsberg N, Barreto M, Placentino A, Dernovsek MZ, Schulze T, Kalember P, Zobel A, Czerski P, Larsen ER, Souery D, Govannini C, Gray JM, Lewis CM, Farmer A, Aitchison KJ, McGuffin P, Craig I. Genetic predictors of antidepressant response in the GENDEP project. Pharmacogenomics J. 2009;9:225–233. doi: 10.1038/tpj.2009.12. [DOI] [PubMed] [Google Scholar]
- 7.Shah RR. Pharmacogenetics in drug regulation: promise, potential and pitfalls. Philos Trans R Soc Lond B. 2005;360:1617–1638. doi: 10.1098/rstb.2005.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomalik-Scharte D, Lazar A, Fuhr U, Kirchheiner J. The clinical role of genetic polymorphisms in drug-metabolizing enzymes. Pharmacogenomics J. 2008;8:4–15. doi: 10.1038/sj.tpj.6500462. [DOI] [PubMed] [Google Scholar]
- 9.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 10.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda T, Yamamoto I, Nishida Y, Zhou Q, Ohno M, Takada K, Azuma J. Effect of the CYP2D6*10 genotype on venlafaxine pharmacokinetics in healthy adult volunteers. Br J Clin Pharmacol. 1999;47:450–453. doi: 10.1046/j.1365-2125.1999.00913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalén P, Dahl ML, Bernal Ruiz MLB, Nordin J, Bertilsson L. 10-Hydroxylation of nortriptyline in white persons with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin Pharmacol Ther. 1998;63:444–452. doi: 10.1016/S0009-9236(98)90040-6. [DOI] [PubMed] [Google Scholar]
- 13.Sachse C, Brockmöller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–295. [PMC free article] [PubMed] [Google Scholar]
- 14.Lessard E, Yessine MA, Hamelin BA, O’Hara G, LeBlanc J, Turgeon J. Influence of CYP2D6 activity on the disposition and cardiovascular toxicity of the antidepressant agent venlafaxine in humans. Pharmacogenetics. 1999;9(4):435–443. [PubMed] [Google Scholar]
- 15.Mulder H, Herder A, Wilmink FW, Tamminga WJ, Belitser SV, Egberts AC. The impact of cytochrome P450-2D6 genotype on the use and interpretation of therapeutic drug monitoring in long-stay patients treated with antidepressant and antipsychotic drugs in daily psychiatric practice. Pharmacoepidemiol Drug Saf. 2006;15:107–114. doi: 10.1002/pds.1173. [DOI] [PubMed] [Google Scholar]
- 16.McAlpine DE, O’Kane DJ, Black JL, Mrazek DA. Cytochrome P450 2D6 genotype variation and venlafaxine dosage. Mayo Clin Proc. 2007;82:1065–1068. doi: 10.4065/82.9.1065. [DOI] [PubMed] [Google Scholar]
- 17.Mulder H, Wilmink FW, Beumer TL, Tamminga WJ, Jedema JN, Egberts AC. The association between cytochrome P450 2D6 genotype and prescription patterns of antipsychotic and antidepressant drugs in hospitalized psychiatric patients: a retrospective follow-up study. J Clin Psychopharmacol. 2005;25:188–191. doi: 10.1097/01.jcp.0000155832.79777.b5. [DOI] [PubMed] [Google Scholar]
- 18.Bijl MJ, Visser LE, Hofman A, Vulto AG, van Gelder T, Stricker BH, van Schaik RH. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br J Clin Pharmacol. 2008;65:558–564. doi: 10.1111/j.1365-2125.2007.03052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy GM, Jr, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry. 2003;160:1830–1835. doi: 10.1176/appi.ajp.160.10.1830. [DOI] [PubMed] [Google Scholar]
- 20.Rudberg I, Hermann M, Refsum H, Molden E. Serum concentrations of sertraline and N-desmethyl sertraline in relation to CYP2C19 genotype in psychiatric patients. Eur J Clin Pharmacol. 2008;64:1181–1188. doi: 10.1007/s00228-008-0533-3. [DOI] [PubMed] [Google Scholar]
- 21.Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther. 2008;83:322–327. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- 22.Yin OQ, Wing YK, Cheung Y, Wang ZJ, Lam SL, Chiu HF, Chow MS. Phenotype-genotype relationship and clinical effects of citalopram in Chinese patients. J Clin Psychopharmacol. 2006;26:367–372. doi: 10.1097/01.jcp.0000227355.54074.14. [DOI] [PubMed] [Google Scholar]
- 23.Fukasawa T, Suzuki A, Otani K. Effects of genetic polymorphism of cytochrome P450 enzymes on the pharmacokinetics of benzodiazepines. J Clin Pharmacol Ther. 2007;32:333–341. doi: 10.1111/j.1365-2710.2007.00829.x. [DOI] [PubMed] [Google Scholar]
- 24.Sim SC, Nordin L, Andersson TM, Virding S, Olsson M, Pedersen NL, Ingelman-Sundberg M. Association between CYP2C19 polymorphism and depressive symptoms. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1160–1166. doi: 10.1002/ajmg.b.31081. [DOI] [PubMed] [Google Scholar]
- 25.Fradette C, Yamaguchi N, Du Souich P. 5-Hydroxytryptamine is biotransformed by CYP2C9, 2C19 and 2B6 to hydroxylamine, which is converted into nitric oxide. Br J Pharmacol. 2004;141:407–414. doi: 10.1038/sj.bjp.0705632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 27.Callen DF, Baker E, Simmers RN, Seshadri R, Roninson IB. Localization of the human multiple drug resistance gene, MDR1, to 7q21.1. Hum Genet. 1987;77:142–144. doi: 10.1007/BF00272381. [DOI] [PubMed] [Google Scholar]
- 28.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 29.Horstmann S, Binder EB. Pharmacogenomics of antidepressant drugs. Pharmacol Ther. 2009;124:57–73. doi: 10.1016/j.pharmthera.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Rochat B, Baumann P, Audus KL. Transport mechanisms for the antidepressant citalopram in brain microvessel endothelium. Brain Res. 1999;831:229–236. doi: 10.1016/s0006-8993(99)01461-4. [DOI] [PubMed] [Google Scholar]
- 31.Crisafulli C, Fabbri C, Porcelli S, Drago A, Spina E, De Ronchi D, Serretti A. Pharmacogenetics of antidepressants. Front Pharmacol. 2011;2:6. doi: 10.3389/fphar.2011.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, Brinkmann U. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakurai A, Onishi Y, Hirano H, Seigneuret M, Obanayama K, Kim G, Liew EL, Sakaeda T, Yoshiura K, Niikawa N, Sakurai M, Ishikawa T. Quantitative structure-activity relationship analysis and molecular dynamics simulation to functionally validate nonsynonymous polymorphisms of human ABC transporter ABCB1 (P-glycoprotein/MDR1) Biochemistry. 2007;46:7678–7693. doi: 10.1021/bi700330b. [DOI] [PubMed] [Google Scholar]
- 34.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 35.Uhr M, Tontsch A, Namendorf C, Ripke S, Lucae S, Ising M, Dose T, Ebinger M, Rosenhagen M, Kohli M, Kloiber S, Salyakina D, Bettecken T, Specht M, Pütz B, Binder EB, Müller-Myhsok B, Holsboer F. Polymorphisms in the drug transporter gene ABCB1 predict antidepressant treatment response in depression. Neuron. 2008;57:203–209. doi: 10.1016/j.neuron.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Peters EJ, Slager SL, Kraft JB, Jenkins GD, Reinalda MS, McGrath PJ, Hamilton SP. Pharmacokinetic genes do not influence response or tolerance to citalopram in the STAR*D sample. PLoS ONE. 2008;3:e1872. doi: 10.1371/journal.pone.0001872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato M, Fukuda T, Serretti A, Wakeno M, Okugawa G, Ikenaga Y, Hosoi Y, Takekita Y, Mandelli L, Azuma J, Kinoshita T. ABCB1 (MDR1) gene polymorphisms are associated with the clinical response to paroxetine in patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:398–404. doi: 10.1016/j.pnpbp.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Vidal R, Pilar-Cuéllar F, dos Anjos S, Linge R, Treceño B, Vargas VI, Rodriguez-Gaztelumendi A, Mostany R, Castro E, Diaz A, Valdizán EM, Pazos A. New strategies in the development of antidepressants: towards the modulation of neuroplasticity pathways. Curr Pharm Des. 2011;17:521–533. doi: 10.2174/138161211795164086. [DOI] [PubMed] [Google Scholar]
- 39.Maron E, Tammiste A, Kallassalu K, Eller T, Vasar V, Nutt DJ, Metspalu A. Serotonin transporter promoter region polymorphisms do not influence treatment response to escitalopram in patients with major depression. Eur Neuropsychopharmacol. 2009;19:451–456. doi: 10.1016/j.euroneuro.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry. 2007;12:247–257. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- 41.Murphy GM, Jr, Hollander SB, Rodrigues HE, Kremer C, Schatzberg AF. Effects of the serotonin transporter gene promoter polymorphism on mirtazapine and paroxetine efficacy and adverse events in geriatric major depression. Arch Gen Psychiatry. 2004;61:1163–1169. doi: 10.1001/archpsyc.61.11.1163. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida K, Takahashi H, Higuchi H, Kamata M, Ito K, Sato K, Naito S, Shimizu T, Itoh K, Inoue K, Suzuki T, Nemeroff CB. Prediction of antidepressant response to milnacipran by norepinephrine transporter gene polymorphisms. Am J Psychiatry. 2004;161:1575–1580. doi: 10.1176/appi.ajp.161.9.1575. [DOI] [PubMed] [Google Scholar]
- 43.Kim H, Lim SW, Kim S, Kim JW, Chang YH, Carroll BJ, Kim DK. Monoamine transporter gene polymorphisms and antidepressant response in Koreans with late-life depression. JAMA. 2006;296:1609–1618. doi: 10.1001/jama.296.13.1609. [DOI] [PubMed] [Google Scholar]
- 44.Higuchi H. Prediction of antidepressant response to milnacipran and fluvoxamine using pharmacogenetical methods. Nihon Shinkei Seishin Yakurigaku Zasshi. 2010;30:71–76. [PubMed] [Google Scholar]
- 45.Min W, Li T, Ma X, Li Z, Yu T, Gao D, Zhang B, Yun Y, Sun X. Monoamine transporter gene polymorphisms affect susceptibility to depression and predict antidepressant response. Psychopharmacology. 2009;205:409–417. doi: 10.1007/s00213-009-1550-3. [DOI] [PubMed] [Google Scholar]
- 46.Dong C, Wong ML, Licinio J. Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Mol Psychiatry. 2009;14:1105–1118. doi: 10.1038/mp.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzeng DS, Chien CC, Lung FW, Yang CY. MAOA gene polymorphisms and response to mirtazapine in major depression. Hum Psychopharmacol. 2009;24:293–300. doi: 10.1002/hup.1024. [DOI] [PubMed] [Google Scholar]
- 48.Lung FW, Tzeng DS, Huang MF, Lee MB. Association of the MAOA promoter uVNTR polymorphism with suicide attempts in patients with major depressive disorder. BMC Med Genet. 2011;12:74. doi: 10.1186/1471-2350-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemonde S, Du L, Bakish D, Hrdina P, Albert PR. Association of the C(-1019)G 5-HT1A functional promoter polymorphism with antidepressant response. Int J Neuropsychopharmacol. 2004;7:501–506. doi: 10.1017/S1461145704004699. [DOI] [PubMed] [Google Scholar]
- 50.Kato M, Fukuda T, Wakeno M, Okugawa G, Takekita Y, Watanabe S, Yamashita M, Hosoi Y, Azuma J, Kinoshita T, Serretti A. Effect of 5-HT1A gene polymorphisms on antidepressant response in major depressive disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:115–123. doi: 10.1002/ajmg.b.30783. [DOI] [PubMed] [Google Scholar]
- 51.Choi MJ, Kang RH, Ham BJ, Jeong HY, Lee MS. Serotonin receptor 2A gene polymorphism (−1438A/G) and short-term treatment response to citalopram. Neuropsychobiology. 2005;52:155–162. doi: 10.1159/000087847. [DOI] [PubMed] [Google Scholar]
- 52.Kato M, Fukuda T, Wakeno M, Fukuda K, Okugawa G, Ikenaga Y, Yamashita M, Takekita Y, Nobuhara K, Azuma J, Kinoshita T. Effects of the serotonin type 2A, 3A and 3B receptor and the serotonin transporter genes on paroxetine and fluvoxamine efficacy and adverse drug reactions in depressed Japanese patients. Neuropsychobiology. 2006;53:186–195. doi: 10.1159/000094727. [DOI] [PubMed] [Google Scholar]
- 53.Minov C, Baghai TC, Schüle C, Zwanzger P, Schwarz MJ, Zill P, Rupprecht R, Bondy B. Serotonin-2A-receptor and -transporter polymorphisms: lack of association in patients with major depression. Neurosci Lett. 2001;303:119–122. doi: 10.1016/s0304-3940(01)01704-9. [DOI] [PubMed] [Google Scholar]
- 54.Lee SH, Lee KJ, Lee HJ, Ham BJ, Ryu SH, Lee MS. Association between the 5-HT6 receptor C267T polymorphism and response to antidepressant treatment in major depressive disorder. Psychiatry Clin Neurosci. 2005;59:140–145. doi: 10.1111/j.1440-1819.2005.01348.x. [DOI] [PubMed] [Google Scholar]
- 55.Illi A, Setälä-Soikkeli E, Viikki M, Poutanen O, Huhtala H, Mononen N, Lehtimäki T, Leinonen E, Kampman O. 5-HTR1A, 5-HTR2A, 5-HTR6, TPH1 and TPH2 polymorphisms and major depression. NeuroReport. 2009;20:1125–1128. doi: 10.1097/WNR.0b013e32832eb708. [DOI] [PubMed] [Google Scholar]
- 56.Perroud N, Aitchison KJ, Uher R, Smith R, Huezo-Diaz P, Marusic A, Maier W, Mors O, Placentino A, Henigsberg N, Rietschel M, Hauser J, Souery D, Kapelski P, Bonvicini C, Zobel A, Jorgensen L, Petrovic A, Kalember P, Schulze TG, Gupta B, Gray J, Lewis CM, Farmer AE, McGuffin P, Craig I. Genetic predictors of increase in suicidal ideation during antidepressant treatment in the GENDEP project. Neuropsychopharmacology. 2009;34:2517–2528. doi: 10.1038/npp.2009.81. [DOI] [PubMed] [Google Scholar]
- 57.Wakeno M, Kato M, Okugawa G, Fukuda T, Hosoi Y, Takekita Y, Yamashita M, Nonen S, Azuma J, Kinoshita T. The alpha 2A-adrenergic receptor gene polymorphism modifies antidepressant responses to milnacipran. J Clin Psychopharmacol. 2008;28:518–524. doi: 10.1097/JCP.0b013e31818455fc. [DOI] [PubMed] [Google Scholar]
- 58.Laje G, Paddock S, Manji H, Rush AJ, Wilson AF, Charney D, McMahon FJ. Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am J Psychiatry. 2007;164:1530–1538. doi: 10.1176/appi.ajp.2007.06122018. [DOI] [PubMed] [Google Scholar]
- 59.Paddock S, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, Lipsky R, Wisniewski SR, Manji H, McMahon FJ. Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. Am J Psychiatry. 2007;164:1181–1188. doi: 10.1176/appi.ajp.2007.06111790. [DOI] [PubMed] [Google Scholar]
- 60.Horstmann S, Lucae S, Menke A, Hennings JM, Ising M, Roeske D, Müller-Myhsok B, Holsboer F, Binder EB. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35:727–740. doi: 10.1038/npp.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Rossum EF, Binder EB, Majer M, Koper JW, Ising M, Modell S, Salyakina D, Lamberts SW, Holsboer F. Polymorphisms of the glucocorticoid receptor gene and major depression. Biol Psychiatry. 2006;59:681–688. doi: 10.1016/j.biopsych.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Wu GS, Luo HR, Dong C, Mastronardi C, Licinio J, Wong ML. Sequence polymorphisms of MC1R gene and their association with depression and antidepressant response. Psychiatr Genet. 2011;21:14–18. doi: 10.1097/YPG.0b013e32834133d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z, Zhu F, Wang G, Xiao Z, Tang J, Liu W, Wang H, Liu H, Wang X, Wu Y, Cao Z, Li W. Association study of corticotropin-releasing hormone receptor1 gene polymorphisms and antidepressant response in major depressive disorders. Neurosci Lett. 2007;414:155–158. doi: 10.1016/j.neulet.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 64.Licinio J, O’Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, Lake S, Tantisira KG, Weiss ST, Wong ML. Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican-Americans. Mol Psychiatry. 2004;9:1075–1082. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 65.Papiol S, Arias B, Gastó C, Gutiérrez B, Catalán R, Fañanás L. Genetic variability at HPA axis in major depression and clinical response to antidepressant treatment. J Affect Disord. 2007;104:83–90. doi: 10.1016/j.jad.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 66.Lin E, Chen PS, Chang HH, Gean PW, Tsai HC, Yang YK, Lu RB. Interaction of serotonin-related genes affects short-term antidepressant response in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:1167–1172. doi: 10.1016/j.pnpbp.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 67.Keers R, Bonvicini C, Scassellati C, Uher R, Placentino A, Giovannini C, Rietschel M, Henigsberg N, Kozel D, Mors O, Maier W, Hauser J, Souery D, Mendlewicz J, Schmäl C, Zobel A, Larsen ER, Szczepankiewicz A, Kovacic Z, Elkin A, Craig I, McGuffin P, Farmer AE, Aitchison KJ, Gennarelli M. Variation in GNB3 predicts response and adverse reactions to antidepressants. J Psychopharmacol. 2011;25:867–874. doi: 10.1177/0269881110376683. [DOI] [PubMed] [Google Scholar]
- 68.Lekman M, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, Lipsky R, Wisniewski SR, Manji H, McMahon FJ, Paddock S. The FKBP5-gene in depression and treatment response–an association study in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Cohort. Biol Psychiatry. 2008;63:1103–1110. doi: 10.1016/j.biopsych.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Appel K, Schwahn C, Mahler J, Schulz A, Spitzer C, Fenske K, Stender J, Barnow S, John U, Teumer A, Biffar R, Nauck M, Völzke H, Freyberger HJ, Grabe HJ. Moderation of adult depression by a polymorphism in the FKBP5 gene and childhood physical abuse in the general population. Neuropsychopharmacology. 2011;36:1982–1991. doi: 10.1038/npp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou YF, Wang Y, Liu P, Feng XL, Wang BY, Zang TH, Yu X, Wei J, Liu ZC, Liu Y, Tao M, Li HC, Li KQ, Hu J, Li M, Zhang KR, Ye DQ, Xu XP. Association of brain-derived neurotrophic factor genetic Val66Met polymorphism with severity of depression, efficacy of fluoxetine and its side effects in Chinese major depressive patients. Int J Neuropsychopharmacol. 2010;13:93–101. doi: 10.1159/000265132. [DOI] [PubMed] [Google Scholar]
- 71.Domschke K, Lawford B, Laje G, Berger K, Young R, Morris P, Deckert J, Arolt V, McMahon FJ, Baune BT. Brain-derived neurotrophic factor (BDNF) gene: no major impact on antidepressant treatment response. Neuropsychobiology. 2010;61:71–78. doi: 10.1017/S1461145709000030. [DOI] [PubMed] [Google Scholar]
- 72.Kocabas NA, Antonijevic I, Faghel C, Forray C, Kasper S, Lecrubier Y, Linotte S, Massat I, Mendlewicz J, Noro M, Montgomery S, Oswald P, Snyder L, Zohar J, Souery D. Brain-derived neurotrophic factor gene polymorphisms: influence on treatment response phenotypes of major depressive disorder. Int Clin Psychopharmacol. 2011;26:1–10. doi: 10.1097/yic.0b013e32833d18f8. [DOI] [PubMed] [Google Scholar]
- 73.Licinio J, Dong C, Wong ML. Novel sequence variations in the brain-derived neurotrophic factor gene and association with major depression and antidepressant treatment response. Arch Gen Psychiatry. 2009;66:488–497. doi: 10.1001/archgenpsychiatry.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heils A, Mossner R, Lesch KP. The human serotonin transporter gene polymorphism—basic research and clinical implications. J Neural Transm. 1997;104:1005–1014. doi: 10.1007/BF01273314. [DOI] [PubMed] [Google Scholar]
- 75.Taylor MJ, Sen S, Bhagwagar Z. Antidepressant response and the serotonin transporter gene-linked polymorphic region. Biol Psychiatry. 2010;68:536–543. doi: 10.1016/j.biopsych.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mrazek DA, Rush AJ, Biernacka JM, O’Kane DJ, Cunningham JM, Wieben ED, Schaid DJ, Drews MS, Courson VL, Snyder KA, Black JL, 3rd, Weinshilboum RM. SLC6A4 variation and citalopram response. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:341–351. doi: 10.1002/ajmg.b.30816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu XZ, Rush AJ, Charney D, Wilson AF, Sorant AJ, Papanicolaou GJ, Fava M, Trivedi MH, Wisniewski SR, Laje G, Paddock S, McMahon FJ, Manji H, Lipsky RH. Association between a functional serotonin transporter promoter polymorphism and citalopram treatment in adult outpatients with major depression. Arch Gen Psychiatry. 2007;64:783–792. doi: 10.1001/archpsyc.64.7.783. [DOI] [PubMed] [Google Scholar]
- 78.Smeraldi E, Serretti A, Artioli P, Lorenzi C, Catalano M. Serotonin transporter gene-linked polymorphic region: possible pharmacogenetic implications of rare variants. Psychiatr Genet. 2006;16:153–158. doi: 10.1097/01.ypg.0000218611.53064.a0. [DOI] [PubMed] [Google Scholar]
- 79.Baffa A, Hohoff C, Baune BT, Muller-Tidow C, Tidow N, Freitag C, Zwanzger P, Deckert J, Arolt V, Domschke K. Norepinephrine and serotonin transporter genes: impact on treatment response in depression. Neuropsychobiology. 2010;62:121–131. doi: 10.1159/000317285. [DOI] [PubMed] [Google Scholar]
- 80.Bonvicini C, Minelli A, Scassellati C, Bortolomasi M, Segala M, Sartori R, Giacopuzzi M, Gennarelli M. Serotonin transporter gene polymorphisms and treatment-resistant depression. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:934–939. doi: 10.1016/j.pnpbp.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 81.Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, Mors O, Placentino A, Rietschel M, Souery D, Zagar T, Czerski PM, Jerman B, Larsen ER, Schulze TG, Zobel A, Cohen-Woods S, Pirlo K, Butler AW, Muglia P, Barnes MR, Lathrop M, Farmer A, Breen G, Aitchison KJ, Craig I, Lewis CM, McGuffin P. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry. 2010;167:555–564. doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 82.Molteni R, Cattaneo A, Calabrese F, Macchi F, Olivier JD, Racagni G, Ellenbroek BA, Gennarelli M, Riva MA. Reduced function of the serotonin transporter is associated with decreased expression of BDNF in rodents as well as in humans. Neurobiol Dis. 2010;37:747–755. doi: 10.1016/j.nbd.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 83.Calabrese F, Molteni R, Cattaneo A, Macchi F, Racagni G, Gennarelli M, Ellenbroek BA, Riva MA. Long-term duloxetine treatment normalizes altered brain-derived neurotrophic factor expression in serotonin transporter knockout rats through the modulation of specific neurotrophin isoforms. Mol Pharmacol. 2010;77:846–853. doi: 10.1124/mol.109.063081. [DOI] [PubMed] [Google Scholar]
- 84.Ham BJ, Lee BC, Paik JW, Kang RH, Choi MJ, Choi IG, Lee MS. Association between the tryptophan hydroxylase-1 gene A218C polymorphism and citalopram antidepressant response in a Korean population. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:104–107. doi: 10.1016/j.pnpbp.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 85.Kato M, Wakeno M, Okugawa G, Fukuda T, Azuma J, Kinoshita T, Serretti A. No association of TPH1 218A/C polymorphism with treatment response and intolerance to SSRIs in Japanese patients with major depression. Neuropsychobiology. 2007;56:167–171. doi: 10.1159/000119734. [DOI] [PubMed] [Google Scholar]
- 86.Secher A, Bukh J, Bock C, Koefoed P, Rasmussen HB, Werge T, Kessing LV, Mellerup E. Antidepressive-drug-induced bodyweight gain is associated with polymorphisms in genes coding for COMT and TPH1. Int Clin Psychopharmacol. 2009;24:199–203. doi: 10.1097/YIC.0b013e32832d6be2. [DOI] [PubMed] [Google Scholar]
- 87.Peters EJ, Slager SL, McGrath PJ, Knowles JA, Hamilton SP. Investigation of serotonin-related genes in antidepressant response. Mol Psychiatry. 2004;9:879–889. doi: 10.1038/sj.mp.4001502. [DOI] [PubMed] [Google Scholar]
- 88.Peters EJ, Slager SL, Jenkins GD, Reinalda MS, Garriock HA, Shyn SI, Kraft JB, McGrath PJ, Hamilton SP. Resequencing of serotonin-related genes and association of tagging SNPs to citalopram response. Pharmacogenet Genomics. 2009;19:1–10. doi: 10.1097/FPC.0b013e3283163ecd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teicher MH, Glod CA, Cole JO. Antidepressant drugs and the emergence of suicidal tendencies. Drug Saf. 1993;8:186–212. doi: 10.2165/00002018-199308030-00002. [DOI] [PubMed] [Google Scholar]
- 90.Healy D, Whitaker C. Antidepressants and suicide: risk–benefit conundrums. J Psychiatry Neurosci. 2003;28:331–337. [PMC free article] [PubMed] [Google Scholar]
- 91.Olfson M, Marcus SC, Shaffer D. Antidepressant drug therapy and suicide in severely depressed children and adults: a case–control study. Arch Gen Psychiatry. 2006;63:865–872. doi: 10.1001/archpsyc.63.8.865. [DOI] [PubMed] [Google Scholar]
- 92.Laje G, Perlis RH, Rush AJ, McMahon FJ. Pharmacogenetics studies in STAR*D: strengths, limitations, and results. Psychiatr Serv. 2009;60:1446–1457. doi: 10.1176/appi.ps.60.11.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased temporal cortex CREB concentrations and antidepressant treatment in major depression. Lancet. 1998;352:1754–1755. doi: 10.1016/S0140-6736(05)79827-5. [DOI] [PubMed] [Google Scholar]
- 94.Dwivedi Y, Rao JS, Rizavi HS, Kotowski J, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Abnormal expression and functional characteristics of cyclic adenosine monophosphate response element binding protein in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60:273–282. doi: 10.1001/archpsyc.60.3.273. [DOI] [PubMed] [Google Scholar]
- 95.Young LG, Bezchlibnyk YB, Chen B, Wang JF, MacQueen GM. Amygdala cyclic adenosine monophosphate response element binding protein phosphorylation in patients with mood disorders: effects of diagnosis, suicide and drug treatment. Biol Psychiatry. 2004;55:570–577. doi: 10.1016/j.biopsych.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 96.Perlis RH, Purcell S, Fava M, Fagerness J, Rush AJ, Trivedi MH, Smoller JW. Association between treatment-emergent suicidal ideation with citalopram and polymorphisms near cyclic adenosine monophosphate response element bind protein in the STAR*D study. Arch Gen Psychiatry. 2007;64:689–697. doi: 10.1001/archpsyc.64.6.689. [DOI] [PubMed] [Google Scholar]
- 97.Dwivedi Y. Brain-derived neurotrophic factor: role in depression and suicide. Neuropsychiatr Disord Treat. 2009;5(433):449. doi: 10.2147/ndt.s5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sairanen M, O’Leary OF, Knuuttila JE, Castren E. Chronic antidepressant treatment selectively increases expression of plasticity related proteins in the hippocampus and medial prefrontal cortex of the rat. Neuroscience. 2007;144:368–374. doi: 10.1016/j.neuroscience.2006.08.069. [DOI] [PubMed] [Google Scholar]
- 99.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- 100.Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 101.Sarchiapone M, Carli V, Roy A, Iacoviello L, Cuomo C, Latella MC, di Giannantonio M, Janiri L, de Gaetano M, Janal MN. Association of polymorphism (Val66Met) of brain-derived neurotrophic factor with suicide attempts in depressed patients. Neuropsychobiology. 2008;57:139–145. doi: 10.1159/000142361. [DOI] [PubMed] [Google Scholar]
- 102.Henningsson S, Borg J, Lundberg J, Bah J, Lindström M, Ryding E, Jovanovic H, Saijo T, Inoue M, Rosén I, Träskman-Bendz L, Farde L, Eriksson E. Genetic variation in brain-derived neurotrophic factor is associated with serotonin transporter but not serotonin-1A receptor availability in men. Biol Psychiatry. 2009;66:477–485. doi: 10.1016/j.biopsych.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 103.Kohli MA, Salyakina D, Pfennig A, Lucae S, Horstmann S, Menke A, Kloiber S, Hennings J, Bradley BB, Ressler KJ, Uhr M, Müller-Myhsok B, Holsboer F, Binder EB. Association of genetic variants in the neurotrophic receptor-encoding gene NTRK2 and a lifetime history of suicide attempts in depressed patients. Arch Gen Psychiatry. 2010;67:348–359. doi: 10.1001/archgenpsychiatry.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng R, Juo SH, Loth JE, Nee J, Iossifov I, Blumenthal R, Sharpe L, Kanyas K, Lerer B, Lilliston B, Smith M, Trautman K, Gilliam TC, Endicott J, Baron M. Genome-wide linkage scan in a large bipolar disorder sample from the National Institute of Mental Health genetics initiative suggests putative loci for bipolar disorder, psychosis, suicide, and panic disorder. Mol Psychiatry. 2006;11:252–260. doi: 10.1038/sj.mp.4001778. [DOI] [PubMed] [Google Scholar]
- 105.Wochnik GM, Rüegg J, Abel GA, Schmidt U, Holsboer F, Rein T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem. 2005;280:4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 106.Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Pütz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Künzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Köhnlein O, Dabitz H, Brückl T, Müller N, Pfister H, Lieb R, Mueller JC, Lõhmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 107.Roy A, Gorodetsky E, Yuan Q, Goldman D, Enoch MA. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology. 2010;35:1674–1683. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Palucha A, Pilc A. The involvement of glutamate in the pathophysiology of depression. Drug News Perspect. 2005;18:262–268. doi: 10.1358/dnp.2005.18.4.908661. [DOI] [PubMed] [Google Scholar]
- 109.Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS ONE. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price RB, Nock MK, Charney DS, Mathew SJ. Effects of intravenous ketamine on explicit and implicit measures of suicidality in treatment-resistant depression. Biol Psychiatry. 2009;66:522–526. doi: 10.1016/j.biopsych.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smits KM, Smits LJ, Schouten JS, Stelma FF, Nelemans P, Prins MH. Influence of SERTPR and STin2 in the serotonin transporter gene on the effect of selective serotonin reuptake inhibitors in depression: a systematic review. Mol Psychiatry. 2004;9:433–441. doi: 10.1038/sj.mp.4001488. [DOI] [PubMed] [Google Scholar]
- 112.de Leon J. The crucial role of the therapeutic window in understanding the clinical relevance of the poor versus the ultrarapid metabolizer phenotypes in subjects taking drugs metabolized by CYP2D6 or CYP2C19. J Clin Psychopharmacol. 2007;27:241–245. doi: 10.1097/JCP.0b013e318058244d. [DOI] [PubMed] [Google Scholar]
- 113.DiazGranados N, Ibrahim LA, Brutsche NE, Ameli R, Henter ID, Luckenbaugh DA, Machado-Vieira R, Zarate CA., Jr Rapid resolution of suicidal ideation after a single infusion of an N-methyl-d-aspartate antagonist in patients with treatment-resistant major depressive disorder. J Clin Psychiatry. 2010;71:1605–1611. doi: 10.4088/JCP.09m05327blu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Jr, Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]