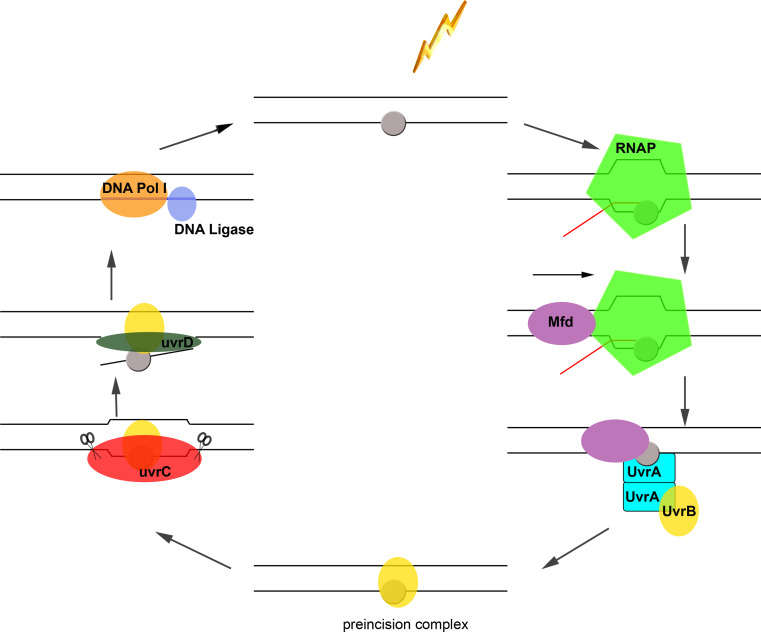
Fig. 1.
Mfd-mediated transcription-coupled DNA repair in bacteria. UV radiation (lightning bolt) induces DNA lesions (grey oval) in the template strand, which induces stalling, and potentially backtracking of elongating RNAP, bound to template DNA (black lines) and nascent transcript (red line). The stalled RNAP complex is recognized by Mfd, which binds the complex from the rear end [67] and subsequently dissociates it after several rounds of ATP hydrolysis [72], during which Mfd tracks and advances on dsDNA [67]. Mfd may then remain bound in proximity to the lesion to recruit the UvrAB complex via direct binding to UvrA. The exact timing of RNAP release within the pathway remains to be established. While the composition of the UvrAB complex recruited during both GG-NER and TC-NER has been a matter of some debate, several recent studies have reported a 2:2 stoichiometry [129, 130]. The presence of two UvrB subunits in the UvrAB complex has been suggested to be required for successive probing of damage of both DNA strands during GG-NER [131]. Due to the high structural similarity and overlapping nature of UvrB/Mfd binding to UvrA [65], it appears likely that Mfd binding may cause dissociation of one UvrB subunit. It is still unknown whether the Mfd-displaced UvrB subunit will eventually form a tight preincision complex with the DNA lesion, or whether this leaves the site so that the second copy of UvrA-bound UvrB can load onto the damaged DNA, as pictured here. UvrC then binds to UvrB to incise the DNA both 3′ and 5′ to the damage, which is followed by removal of the oligonucleotide by UvrD, gap filling and ligation by DNA polymerase I and DNA ligase [62]. The relative sizes of the nucleic acids and proteins depicted schematically do not reflect relative sizes or conformations