Fig. 3.
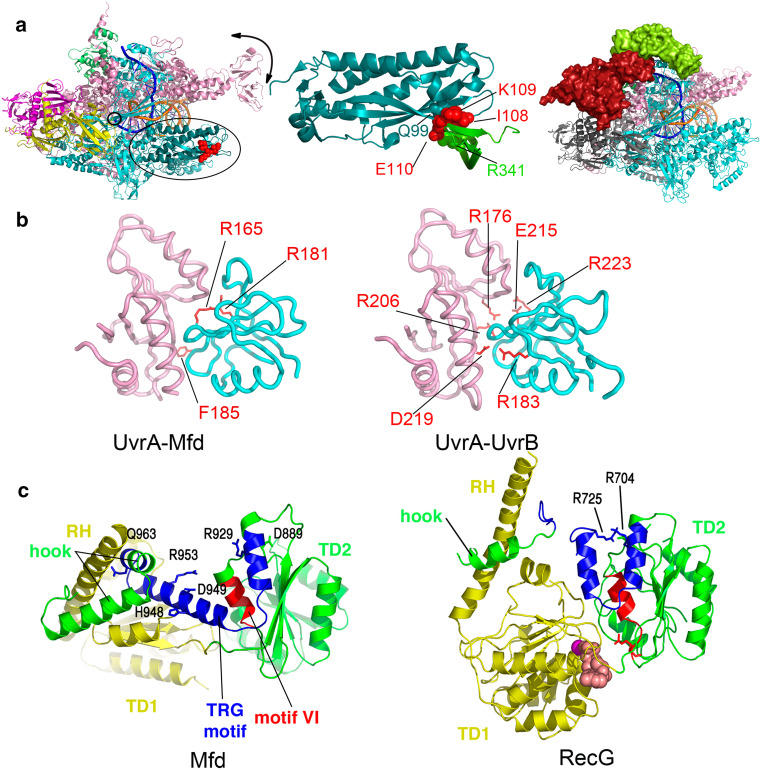
Binding modules within the bacterial TRCFs, Mfd, and Nus. a Binding of the bacterial transcription-repair coupling factors Mfd and NusA to RNAP. Overall architecture of Thermus thermophilus RNAP elongation complex (PDB ID 2O5I) is shown on the left (with RNAP subunits colored as follows: α1 and α2 in magenta and yellow, β in cyan, β’ in pink, ω in green) with the N-terminal region of the β subunit interacting with Mfd-RID colored in deep teal and indicated by an ellipse. Downstream DNA is shown as going into the page, the catalytic center is indicated by a small circle, and movement of the RNAP clamp is indicated schematically by a black arrow. The crystallographic model of Thermus sp. core Mfd-RID/RNAP-β complex (PDB ID 3MLQ) [82] is shown in the middle, while the pseudoatomic model of Bacillus subtilis NusA bound to a TEC, derived from single-particle electron microscopy studies and homology modeling [108], is shown on the right, with the NusA-NTD colored in lime and NusA-CTD, positioned lower down the β flap towards the α-subunit, colored in red. b Mfd and UvrB share a similar mode of binding to UvrA. Shown are core E. coli Mfd-UvrA (left, PDB ID 4DFC) and Geobacillus stearothermophilus UvrA-UvrB (right, PDB ID 3FPN) complexes, with residues important for the respective interaction [79, 132] highlighted as red sticks. Mfd-D2 domain (and the homologous UvrB domain) is colored cyan, and UvrA is shown in pink. c dsDNA translocation and the TRG motif in the Mfd and RecG proteins. Side-by-side views of the translocation modules of nucleotide-free E. coli Mfd (left, PDB ID 2EYQ) and ADP-bound Thermotoga maritima RecG (right, PDB ID 1GM5), illustrating the conformational changes in the relay helix, helicase motif VI (red), TRG motif (blue), and the hook. The two views are aligned such that translocase domain 2 (TD2 or D6 in Mfd, colored green throughout) of each molecule is in identical orientation. Side chains of key residues are shown. Reprinted with permission from [61]