Fig. 1.
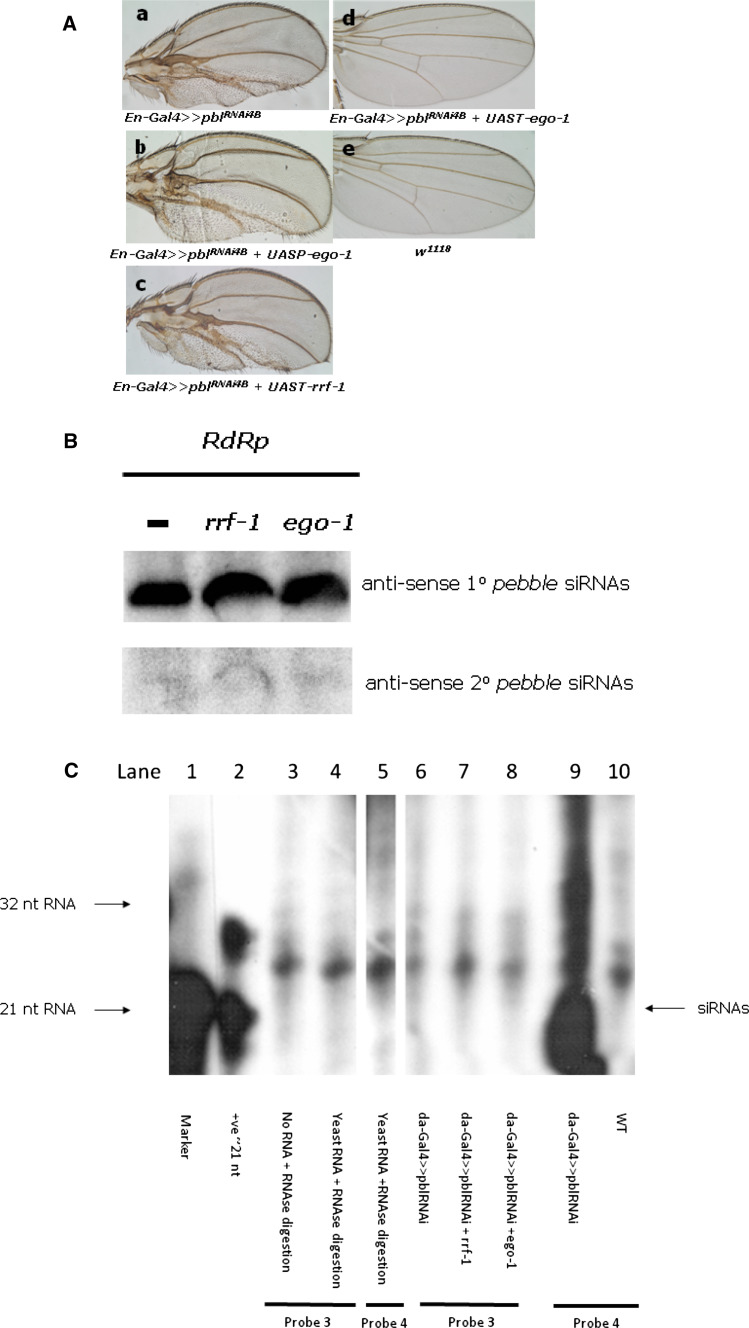
Silencing of the endogenous gene, pebble, in Drosophila and detection of the primary as well as secondary siRNAs. a Expression of canonical C. elegans RdRps does not noticeably enhance RNAi-mediated silencing of the Drosophila endogenous gene, pebble. For activation of the dsRNA expression, flies harboring the pebble gene dsRNA construct pblRNAi 4B(3.1) were crossed with the tissue-specific driver en-Gal4 in the presence (b–d) or absence (a) of one of the RdRps, RRF-1 or EGO-1. Compared with the control (a), noticeable changes in pebble gene silencing in either b or c were not observed; however, expression of higher levels of EGO-1 from the UAST vector gave a wild-type phenotype (d) through comparison with WT (e). b Northern hybridization did not detect any production of secondary siRNAs. Probe 1, designed based on the pebble targeted sequence (Fig. S1), successfully detected 1° siRNAs, while probe 2, designed based on the sequence upstream of the targeted sequence, did not detect any 2° siRNA production regardless of the presence of either of the RdRp genes. We used the RNA samples from UASP-ego-1 rather than UAST-ego-1 as da-gal4-activated UAST-ego-1 automatically silenced pblRNAi 4B(3.1) and gave WT phenotype, i.e., could not suspend the development at pupae stage as occurred in UAST-rrf-1 and UASP-ego-1. c Secondary siRNAs detection with RNAse protection assays. RNAs was prepared from pupae. Lane 1: RNA marker, lane 2: positive control RNA; lanes 3 and 4 are controls for probe 3 with yeast or no RNA; lane 5 is control for probe 4 with RNAse digestion. Lanes 6–8 are the results for detection of 2° siRNAs upstream of the targeted pebble sequence (Fig. S1 and S2) with probe 3; da-Gal4-activated rrf-1 (lane 7) or ego-1 (lane 8) did not generate detectable 2° siRNAs. Lane 9: detection of the primary siRNAs with probe 4, lane 10: no detection in w 1118