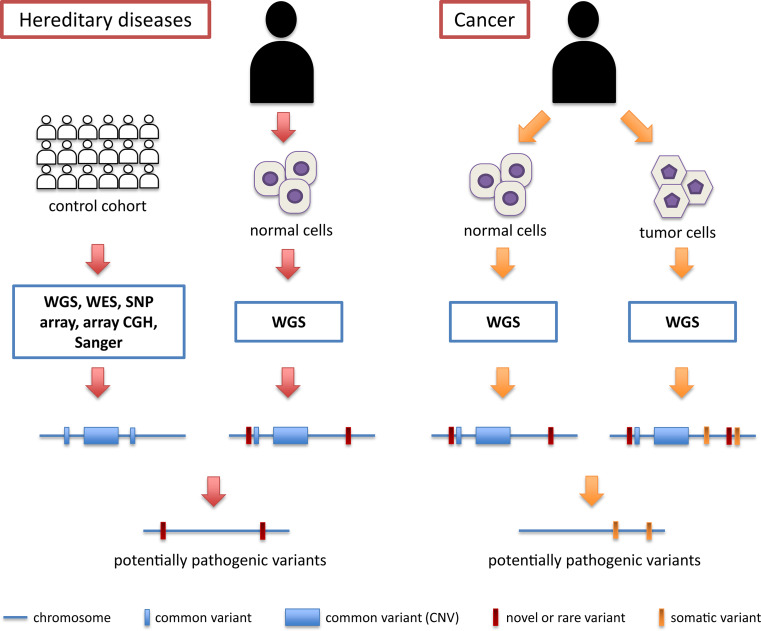
Fig. 1.
Schematic workflow for the detection of potentially pathogenic DNA variants in hereditary diseases and in cancer. In hereditary diseases, the information from several genomes from a control cohort (white individuals) is assembled to produce a “metagenome” that includes all possible variants (both small events and copy number variations, or CNVs) that are allegedly not causing disease in the general population (blue bars and boxes). Potentially pathogenic variants are then deduced by comparing the WGS information from a patient (black individual) with that of the metagenome. In cancer, there is no need to query a control cohort, since the control information is provided by the genome of normal cells from the same patient. Regardless of their frequency in the general population, all these variants are then subtracted from the pool of DNA changes obtained from the tumor genome, making the detection process of pathogenic variants a more efficient and straightforward procedure