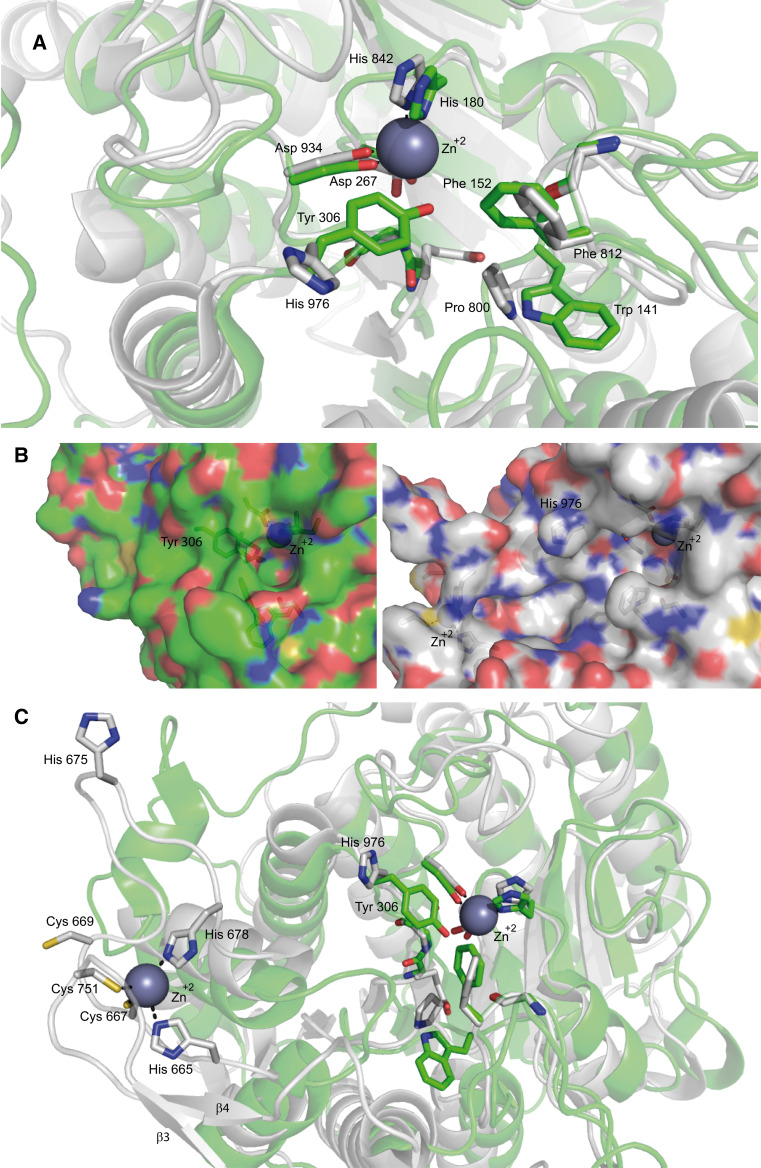
Fig. 2.
Representation of class I and class IIa catalytic sites (a, b) and the zinc binding domain (c). a Superimposition of the inhibitor (TFMK)-bound ribbon structure of class I HDAC8 (green) and of class IIa HDAC4 (white) catalytic sites. As mentioned in the text the His 976 is rotated away from the active site differently from Tyr 308 in HDAC8. b Surface representation of class I HDAC8 (green) and class IIa HDAC4 (white) catalytic sites. The figure shows the hydrophilic tunnel necessary for the release of the reaction product in HDAC8 (green), while in HDAC4 (white) the His/Tyr substitution prevents tunnel formation. c Superimposition of the inhibitor (TFMK)-bound ribbon structure of class I HDAC8 (green) and of class IIa HDAC4 (white) catalytic site (right) and zinc binding domain of HDAC4 (left). β3 and β4 are the two antiparallel β-strands involved in the formation of the pocket-like structure in the zinc binding domain. Importantly, His 665 and His 678 in this inhibitor-bound structure are replaced by Cys 669 and His 675 in the coordination of the zinc ion in the Apo-structure. Unfortunately the crystallization of Apo-HDAC4 was unsuccessful and these differences are deduced from crystallographic studies of the mutant GOF (H976Y) of HDAC4 [31]. The coordinates of the protein structures were retrieved from the protein data bank. Amino acids discussed in the text are labeled and shown in stick representation. The accession codes for the protein structures are: 2VQJ (HDAC4) and 1T69 (HDAC8). Figures are edited using PyMOL Molecular graphics system, Schrödinger, LLC