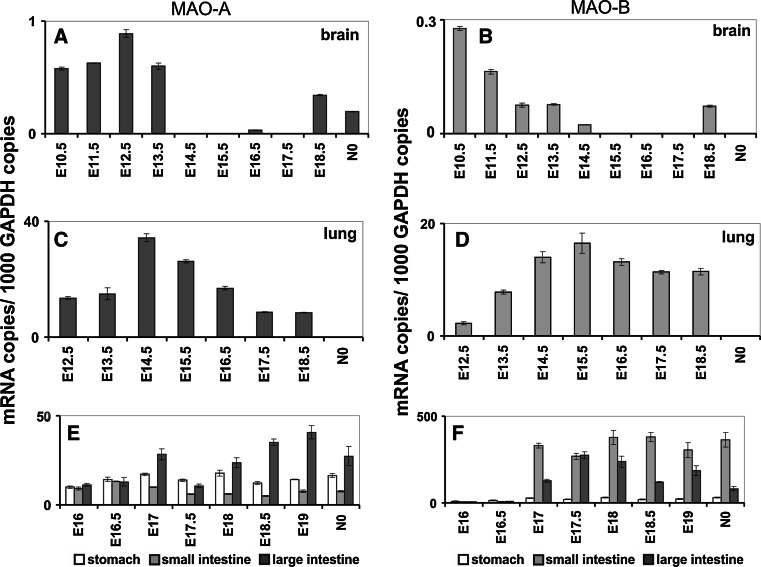
Fig. 6.
Expression of MAO-A and MAO-B in the embryonic brain (a, b), lung (c, d) and gastrointestinal tract (e, f). Inbred ICR pregnant mice were obtained from the animal house, and embryos at different developmental stages [gestational day 6.5 (E6.5) to E17.5] were prepared under a stereomicroscope (Olympus, USA). For qRT-PCR, preparations were kept in PBS (0.1 % diethyl pyrocarbonate), and extra-embryonic tissue was removed. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Germany) and was reversely transcribed according to standard protocols with oligo d(T)15 primers and SuperScript III reverse transcriptase (Invitrogen) according to the vendor’s instructions. Quantitative RT-PCR (RT-qPCR) was carried out with a Rotor Gene 3000 system (Corbett Research, Australia) using ImmoMix/SYBR Green (BIOLINE, Germany). Isoform-specific amplification primers were designed for MAO-A (5′-TTC AGC GTC TTC CAA TGG GAG CT-3′/5′-TGC TCC TCA CAC CAG TTC TTC TC-3′), MAO-B (5′-ACT CGT GTG CCT TTG GGT TCA G-3′/5′-TGC TCC TCA CAC CAG TTC TTC TC-3′) and GAPDH (5′-CCA TCA CCA TCT TCC AGG AGC GA-3′/5′-GGA TGA CCT TGC CCA CAG CCT TG-3′). Absence of cross-amplification between the two isoforms was ensured, and the standard protocol was followed as outlined before [296]. RNA preparations were analyzed at least in triplicate, and mean ± SD are given. The experimental raw data were evaluated with the Rotor-Gene Monitor software (version 4.6). To generate standard curves for quantification of expression levels, specific amplicons were used as external standards for each target gene (5 × 103–3 × 106 copy numbers). GAPDH mRNA was used as internal standard to normalize expression of the target transcripts