Fig. 3.
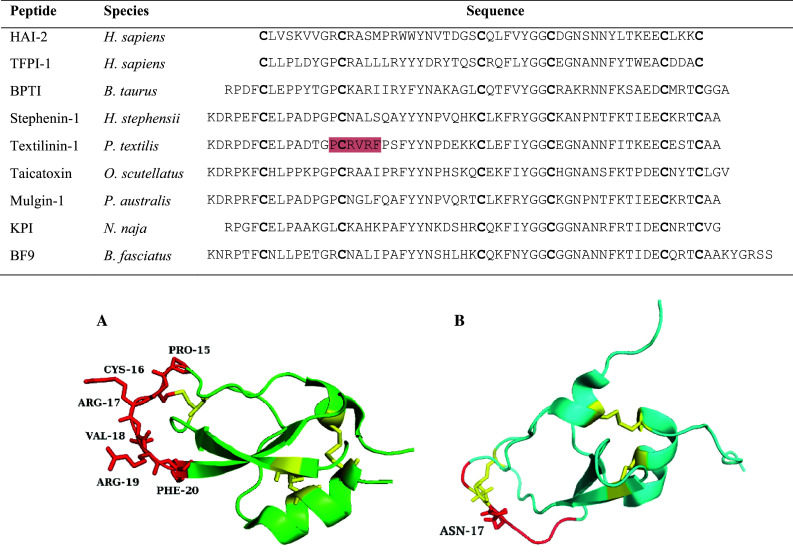
Kunitz-type serine protease inhibitor sequence alignment. The residues highlighted in red on the textilinin-1 sequence are the amino acids involved in the anatagonistic interaction with the protease active site. Conserved cysteines are shown in bold. Accession numbers are as follows: O43291, P10646, P00974, B5KF94, Q90WA1, B7S4N9, Q6ITC1, P19859, B2KTG1. 3D structures are shown for A Textilinin-1 (3BYB) and B chymotrypsin inhibitor from Bungarus fasciatus (1JC6). Textilinin-1 shows the P3–P3′ residues in red with Arg17 (P1′) typical for trypsin inhibition. The chymotrypsin inhibitor denotes the P3–P3′ residues in red but with an asparagine residue at P1′ instead of the typical hydrophobic residues of Leu, Phe and Tyr. Disulfide bridges are depicted in yellow
