Abstract
Trichomes, originating from epidermal cells, are present on nearly all terrestrial plants. They exist in diverse forms, are readily accessible, and serve as an excellent model system for analyzing the molecular mechanisms in plant cell differentiation, including cell fate choices, cell cycle control, and cell morphogenesis. In Arabidopsis, two regulatory models have been identified that function in parallel in trichome formation; the activator–inhibitor model and the activator–depletion model. Cotton fiber, a similar unicellular structure, is controlled by some functional homologues of Arabidopsis trichome-patterning genes. Multicellular trichomes, as in tobacco and tomato, may form through a distinct pathway from unicellular trichomes. Recent research has shown that cell cycle control participates in trichome formation. In this review, we summarize the molecular mechanisms involved in the formation of unicellular and multicellular trichomes, and discuss the integration of the cell cycle in its initiation and morphogenesis.
Keywords: Trichome, Differentiation, Cell cycle, Cell fate determination
Introduction
Trichomes are easily accessible appendages originating from the aerial epidermal cells of leaves, stems, and floral organs in plants. Although not essential for plant growth, they serve a variety of important functions, e.g., protection against insect and pathogen attack, reducing water loss, and increasing tolerance to abiotic stress conditions, including extreme temperatures and UV irradiation [1–5]. Furthermore, glandular trichomes provide chemical protection by synthesizing and secreting specialized metabolites [6–8]. There are many types of trichomes characterized by their morphology, location, and nature; they may be unicellular or multicellular, branched or non-branched, glandular or non-glandular. Although trichomes vary considerably in morphology, the cells destined to become trichomes must undergo similar transition from the mitotic cell cycle to an endoreduplication cycle [9]. In Arabidopsis, trichomes are typically unicellular structures and their formation in the epidermis involves three different stages: cell fate determination, cellular specification, and morphogenesis [10]. The cells forming trichomes are specified, and while the other epidermal cells continue to divide, trichome cells enter a phase of four endoreduplication cycles, reaching an average DNA content of 32C [11]. In contrast, multicellular trichomes in tobacco and tomato are formed by a change in the growth orientation of epidermal cells, which divide and grow perpendicular to the leaf surface [12].
In the last two decades, molecular mechanisms involved in the initiation and development of trichomes have been investigated with the objective of analyzing factors controlling cell fate and differentiation in plant cells. With the availability of a large number of trichome-related mutants in Arabidopsis, many key regulators controlling trichome formation have been identified. The GLABROUS1 (GL1) gene, which encodes an R2R3 MYB transcription factor, was the first to be cloned [13, 14], and a MYB/bHLH/WD-repeat complex consisting of GL1, TRANSPARENT TESTA GLABRA1 (TTG1), GLABRA3 (GL3), and ENHANCER OF GLABRA3 (EGL3) was determined, providing significant insight into the genetic control of unicellular trichome formation [15–18]. These regulators also shed light on the regulation of cotton fiber formation. For instance, GaMYB2, a homologue of GL1 in cotton, can induce fiber initiation in cotton and trichome formation in Arabidopsis, respectively [19]. However, our knowledge of the regulatory mechanisms in multicellular trichome formation is much less known. Different developmental events in unicellular and multicellular trichomes suggest that the latter are likely controlled by different molecular mechanisms. Moreover, genetic studies in tobacco, which produce multicellular trichomes, support the view that different types of trichomes are induced by distinct pathways [20, 21].
In recent years, it has become clear that cell cycle regulation plays a pivotal role in the differentiation of plant structures. During trichome formation, many important regulators participating in cell cycle control have been cloned and characterized, e.g., SIAMESE (SIM), TRIPTYCHON (TRY), SlCycB2, and the cell cycle-related factors [22–24]. This review summarizes the research progress on the molecular mechanisms in unicellular and multicellular trichome formation, and the regulation of the cell cycle in its initiation and morphogenesis.
Arabidopsis trichomes: a model of trichome development and regulation
In Arabidopsis, trichomes are produced on nearly all the aerial organs except for the hypocotyl and the cotyledons [25]. Trichomes on rosette leaves are large single cells, usually with three branches, and until now have been the subject of research. The initiation of trichomes occurs at the base of young leaves, in which all cells are potentially equivalent to develop into trichomes [26], but trichomes are spaced at a regular interval of three or four cells from each other [10]. Trichome clusters are never observed in leaf epidermis, implying that there must be a mechanism regulating the spacing pattern. It is hypothesized that the establishment of the trichome pattern is based on the activator–inhibitor mechanism: initial epidermal cells equivalently express trichome-promoting factors, which can activate repressors but result in different cell fate in the neighboring cells [27, 28]. Numerous trichome-related mutants have enabled the identification of many trichome-patterning genes, which can be divided into two types: trichome-positive and trichome-negative regulators. The positive regulators include the R2R3 MYB transcription factor GL1 and its functionally equivalent counterparts WEREWOLF (WER) and MYB23, the bHLH factor GL3 and its close homolog EGL3 and the WD40-repeat factor TTG1 [14, 16, 18, 29, 30]. GL1 and TTG1 interact with GL3/EGL3, respectively, forming a MYB/bHLH/WD-repeat complex [15] (Fig. 1). This regulatory complex stimulates epidermal cells to differentiate into trichomes by activating the expression of its downstream activators GLABRA2 (GL2) and TRANSPARENT TESTA GLABRA2 (TTG2), which encodes a homeodomain-leucine zipper (HD-Zip) and a WRKY transcription factor individually [31–34] (Fig. 1). The negative regulators are represented by six redundantly acting genes: CAPRICE (CPC), TRY, ENHANCER OF TRY AND CPC 1 (ETC1), ETC2, ETC3 and TRICHOMELESS1 (TCL1), all of which encode single-repeat R3 MYB proteins [23, 35–40] (Fig. 1). These small-sized inhibitors can move laterally into neighboring cells and compete with GL1 for binding to GL3/EGL3 [41], forming an inactivating complex, which cannot promote GL2 and TTG2 expression, thereby inhibiting trichome fate [15].
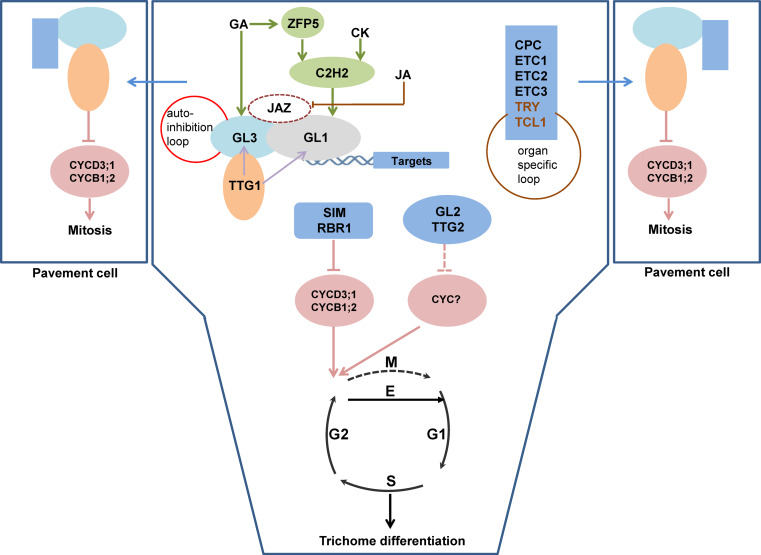
Fig. 1.
Model of trichome cell fate determination in Arabidopsis. GL3 physically interacts with GL1 and TTG1 separately, forming a MYB/bHLH/WD-repeat complex. Three phytohormones (GA, CK, and JA) participate in the control of trichome initiation. GA activates the expression of ZFP5, and then ZFP5 induces GIS (C2H2) expression (green arrows). In addition, ZFP8 (C2H2) and GIS2 (C2H2) are simultaneously activated by ZFP5 and CK (green arrows). The transcription of GL1 is enhanced by C2H2. JA regulates trichome formation by degrading JAZ proteins and then abolishing the interactions of JAZ proteins with bHLH and MYB factors (brown inhibitory line). The MYB/bHLH/WD-repeat complex stimulates trichome formation by activating the expression of its direct targets (SIM and RBR1 as cell cycle regulators, GL2 and TTG2 as transcription factors, and CPC, TRY, ETC1, ETC2, ETC3, and TCL1 as inhibitors). SIM and RBR1 induce trichome differentiation by repressing the expression of cell cycle-related genes, CYCD3;1 and CYCB1;2, which triggers the transition of mitosis to endoreduplication cycles. How GL2 and TTG2 regulate trichome initiation remains uncharacterized. Maybe they also control the expression of cell cycle-related genes (CYC). Inhibitors (represented by the blue rectangle) move to the neighboring cells and substitute GL1 in the complex, resulting in the loss of the activation activity of CYCD3;1 and CYCB1;2 and then leading to mitosis. Note, TTG1 acts upstream of GL3 and GL1, and can activate their transcription (purple arrows); an auto-inhibition was observed for GL3, which can bind to its own promoter (red circle); TRY and TCL1 also participate in the organ-specific control of trichome formation (brown circle). Five phases during cell cycle: M (mitosis), G1 (gap 1), S (synthesis), and G2 (gap 2), endoreduplication (E)
Analysis of the transcriptional regulation of these patterning genes is very important for unfolding the underlying mechanisms of trichome formation. It was reported that GL1 and GL3 contain a DNA-binding domain, respectively [42], and deletion of the DNA-binding domains completely represses the expression of their downstream gene GL2, suggesting that GL2 may be positively regulated by them through DNA–protein binding [42]. Thereafter, it was suggested that GL1 directly binds to the promoters of GL2, TTG2, CPC, ETC1, and ETC3, which corresponds to the finding that TTG2 is directly regulated by GL1 [15, 32, 43] (Fig. 1). Simultaneously, both TTG1 and GL3 were demonstrated to interact with the promoters of TTG2, CPC, and ETC1 [15, 31]. The activation of CPL3 and TRY promoters requires direct binding of GL3 [44] (Fig. 1). More importantly, using the chromatin immunoprecipitation (ChIP)-chip method, some novel direct targets of GL1 and GL3 were determined, i.e., MYC1 and SCL8 acting as transcription factors, and SIM and RETINOBLASTOMA RELATED1 (RBR1), which are involved in cell cycle regulation [44] (Fig. 1). These regulatory relationships suggest that these factors are major downstream targets for the MYB/bHLH/WD-repeat complex. Compared with the double-mutants gl2 and single myb repressors, the glabrous phenotype in the gl2 single mutants was partially rescued, indicating single MYB genes may not act solely via GL2 [45]. SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) promotes TCL1 and TRY expression by direct binding with their promoters, which participate in temporal control of trichome differentiation [46]. In addition to these relationships, the interaction network between the members of the MYB/bHLH/WD-repeat complex was also investigated in some studies. Neither GL1 rescued the phenotype of gl3 mutant, nor GL3 the phenotype of gl1 mutant, suggesting that these two genes function at the same hierarchical level [47, 48]. TTG1 can directly activate the GL3/GL1 transcription, indicating that TTG1 may act upstream of GL3 and GL1 [15] (Fig. 1). Moreover, GL3 was found to bind to its own promoter and an auto-inhibition was shown for this gene using co-transfection assays in protoplasts [31] (Fig. 1). These findings further our understanding of the activator–inhibitor mechanism of trichome formation.
The model described above cannot explain the paradoxical genetic evidence that strong TTG1 alleles exhibit glabrous, whereas weak alleles trichome clusters phenotypes, suggesting this gene functions as both a negative and positive regulator [49]. The TTG1 protein has been shown to move between epidermal cells of young Arabidopsis leaves and bind to GL3 [26, 49]. Initially, all cells ubiquitously express TTG1, whereas the expression level was dramatically decreased in the cells adjacent to the developing trichomes. Consequently, cells with high GL3/TTG1 levels are more competent, whereas their neighbors lacking TTG1 are less competent for trichome cell fate determination [26]. Thus, an activator–depletion model for trichome patterning was conceived based on the GL3-dependent depletion of TTG1 in trichome neighboring cells, and it may function in parallel to the activator–inhibitor mechanism [49]. Balkunde et al. [50] further studied the molecular basis of this trapping mechanism and found that TTG1 mobility was regulated by GL3, and that the trapping in different tissue layers depended on direct interaction between them and recruitment in the nucleus.
Trichome formation is clearly a complicated but a precisely regulated biological process, which cannot be fully explained by the two models discussed above. Moreover, some observations pose new challenges and demand further explanations. For example, it remains unclear why the mutation in TRY results in trichome clusters and other inhibitors participate in trichome density control, and it also remains unexplained which signals determine the movement direction of these regulators between epidermal cells. Furthermore, it is still not known how organ-specific differences in trichome formation are achieved. Fortunately, new findings shed additional light on trichome-patterning mechanisms. Pesch and Hülskamp [51] analyzed the transcriptional regulation of TRY and identified a cis-element in its promoter region required for the repression of TRY auto-repression, revealing specific properties of this gene for clustering regulation (Fig. 1). Wang et al. [40] demonstrated that overexpression of TCL1 can completely suppress trichome initiation on all aerial organisms, whereas loss-of-function mutation in this gene activates unique, ectopic trichome formation on the inflorescence stems and pedicels. Interestingly, the expression of GL1 is directly suppressed by TCL1 through binding with its promoter, indicating a novel regulatory loop and a new insight into the organ-specific control in trichome patterning (Fig. 1) [40]. TCL2, which encodes a previously uncharacterized single repeat MYB protein exhibiting 80 % amino acid identities with TCL1, negatively regulates trichome formation like other single-repeat MYBs [52]. Ectopic expression of TCL2 under the control of TCL1 promoter did not fully rescue the mutant phenotype of tcl1, suggesting that these two genes do not function equivalently [52]. 1-Deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) plays an important role in trichome formation in Arabidopsis [53], as the dxr mutant and co-suppression of transgenic lines produced much less trichomes. It was pointed out that DXR is a key enzyme involved in the 2-C-methyl-derythritol-4-phosphate (MEP) pathway [53]. Further studies showed that trichome initiation-related genes (GL1, TTG1, TRY, and SPINDLY) and phytohormones gibberellins (GAs) were also affected in dxr mutant [53] and that GA was synthesized from the plastidial geranyl-geranyl diphosphate precursor in the MEP pathway. We thus inferred that DXR affects trichome formation by regulating the synthesis of GA, which controls trichome-patterning gene expression. Recently, an amino acid substitution in both ETC2 and AtMYC1, respectively, was identified, and was shown to be responsible for trichome density variation in natural populations [54, 55]. This suggests that much remains unknown in the coding sequences of the patterning genes, which may not be characterized merely by overexpression and suppression of regulators. Surprisingly, new experiments have revealed that epigenetic factors are also associated with trichome density. In Mimulus guttatus, parental leaf damage can induce increased trichome density in progeny by down-regulating the expression of MYB MIXTA-like 8, which is correlated with an epigenetic mechanism [56]. It would be interesting to determine whether there is epigenetic regulation of trichome-patterning genes. Kryvych et al. [57] and Lieckfeldt et al. [58] analyzed the gene-expression profiles of Arabidopsis trichomes and identified some novel genes involved in their initiation and development, indicating that comprehensive transcriptional studies should help us unfold new regulators of trichome formation.
Phytohormones, e.g., GAs, cytokinins (CKs), and jasmonates (JAs), play important roles in plant growth and development. However, not much is known of their roles in the regulation of trichome formation. Application of GA was first reported to stimulate trichome formation in the glabrous GA-deficient mutant gal-3 [59, 60] and the GA content was positively correlated with trichome branch number in Arabidopsis [61]. Exogenous CK causes trichome proliferation on the inflorescence stem, indicating that CK also participates in trichome initiation [46]. However, stimulation of trichome initiation by GA is inhibited by CK application, demonstrating important interactions between them in trichome formation [62]. JA and its derivative compounds function as key signaling molecules in trichome formation and exogenous JA causes an increase in the number of leaf trichomes in Arabidopsis [63]. Interestingly, GA and JA have a synergistic effect on trichome induction. Thus, these three phytohormones positively regulate trichome initiation, whereas salicylic acid had a negative effect on trichome production [63]. Despite these relationships, the molecular mechanisms by which phytohormones control trichome formation still remain elusive. It would be useful to determine whether the effects of these phytohormones are mediated by regulating the expression of the essential components of MYB/bHLH/WD-repeat transcriptional complex. GL1 was the first gene which was shown to be up-regulated by GA [61]; its transcription was repressed in GA-deficient mutant, but activated by exogenous GA [64]. Moreover, the transcription levels of GL3, TTG1, and TRY were also regulated by GA. Both GA and CK are integrated by GLABROUS INFLORESCENCE STEMS (GIS), GIS2, and ZINC FINGER PROTEIN 8 (ZFP8), all of which encode the C2H2 zinc-finger transcription factors with equivalent functions, and they collectively control GL1 transcription [65]. A new zinc-finger protein, ZFP5, also acts as an activator in controlling trichome production through GA signaling, which functions upstream of GIS, GIS2, ZFP8, GL1, and GL3 [66] (Fig. 1). In Arabidopsis, jasmonate ZIM-domain proteins repress trichome formation by binding with GL3, EGL3, and GL1, key partners of the activation complex [67]. Also, JA participates in trichome initiation by degrading JAZ proteins and then abolishing the interactions of JAZ proteins with bHLH and MYB factors, which activate the transcription of trichome activators [68] (Fig. 1). In addition, GL3 coordinately promotes trichome cell fate in response to JA with GL1 [69]. These new findings extend our knowledge of the molecular mechanisms in the phytohormone regulation of trichome formation.
Cotton fiber: another type of unicellular trichome and its regulation
Cotton fibers, a useful material for the textile industry, are single-celled structures originating from the ovule epidermis. Cotton fibers elongate via a linear growth model [70], their developmental process consists of four overlapping stages: initiation, elongation, secondary wall deposition, and maturation. Molecular investigations of cotton fiber formation have provided valuable data on our understanding of plant cell fate. Several transcription factors are expressed in developing fibers, some of which display high sequence identities with Arabidopsis trichome pattering genes. For example, two R2R3 MYB genes GhMYB1 and GaMYB2 with high sequence similarities to GL1, control fiber cell determination [19, 70]. Ectopic expression of either GhMYB1 or GaMYB2 under control of the GL1 promoter can restore trichome formation in gl1 mutant [19, 20]. The downstream gene of GaMYB2, cotton fiber-related GhRDL, could activate trichome initiation in Arabidopsis seed [71]. Two other MYB regulators, GhMYB25 and GhMYB25-like, have been reported, and suppression of their expression in cotton results in reduction of fiber number [72, 73]. An L1 box binding protein, GbML1, interacting with GbMYB25, was also identified to activate cotton fiber initiation [74]. Moreover, two cotton genes homologous to Arabidopsis TTG1 rescue trichome phenotype when expressed in the ttg1 mutant of Arabidopsis, indicating that they are functional homologues of TTG1 [75]. Functional homologues of GL2 have also been reported in cotton. Three HD-ZIP transcriptional factors (GaHOX1, GaHOX3 and GaHOX3) were cloned and characterized in cotton and GaHOX1 could functionally substitute GL2 when introduced into Arabidopsis [76]. Although no homologues of GL3 or EGL3 have been identified in cotton fiber initiation, we speculate a transcriptional complex similar to the MYB/bHLH/WD-repeat complex in Arabidopsis in cotton. However, whether the expression of GaHOX1 is regulated by the putative activating complex remains unknown. Unlike Arabidopsis, the negative regulators like single-repeat R3 MYB transcription factors have not been identified in cotton, and it is unknown whether fiber cell fate depends on the activator–inhibitor mechanism. Cotton PROTODERMAL FACTOR 1 (GbPDF1) is exclusively expressed in the L1 layer of many tissues, encoding a putative extracellular proline-rich protein [75], and GbPDF1 together with interaction partners functions in cotton fiber initiation through its core cis-element HDZIP2ATATHB2 [77]. Further research is needed to fully understand the molecular mechanisms in cotton fiber formation.
Multicellular trichome formation and its regulation
The majority of flowering plants produce multicellular trichomes, which undergo three processes: initial commitment, expansion, and morphogenesis, with some similarity to unicellular trichomes. The epidermal cells destined to become multicellular trichomes first enlarge, and then divide perpendicular to the epidermal surface with continued cell division [78] (Fig. 2). However, there is no experimental evidence documenting the transition from mitosis to endoreduplication in multicellular trichomes, and little is known of the molecular mechanisms in their initiation and development. Tobacco (Nicotiana tabacum) produces short- and long-stalked multicellular trichomes, and these two types of trichomes are produced by different developmental programs [20]. MIXTA, an R2R3-type MYB transcription factor controlling petal conical cell formation in snapdragon (Antirrhinum majus), could trigger long-stalked trichome formation when ectopically expressed in tobacco [20], suggesting that conical cells and multicellular trichomes share some common elements. Two homologues of MIXTA, MYB MIXTA LIKE 1 (AmMYBML1) from snapdragon, and CotMYBA from cotton could also promote multicellular trichome formation when overexpressed in tobacco [20, 79]. These results suggested that unidentified MIXTA-like genes inducing multicellular trichome formation likely exist in tobacco. Interestingly, two MIXTA-like genes activating trichome differentiation in woody nightshade (Solanum dulcamara) and petunia (Petunia hybrida) have been characterized [21, 80]. As these R2R3 MYB-related transcriptional regulators contain the conserved DNA-binding domain similar to that of GL1, we presume that the molecular basis of trichome formation in these species is similar to that in Arabidopsis and cotton. However, over-expression of GL1 in tobacco had no effect on trichome formation [20]. In addition, ectopic expression of MIXTA in Arabidopsis gl1-1 mutant failed to restore the trichome phenotype [20]. These data, together with the absence of amino acid signature in MIXTA and MIXTA-like proteins required for the interaction with GL3 and EGL3, suggest that trichome formation in tobacco may not be controlled by the MYB/bHLH/WD-repeat complex [81]. This view is supported by the finding that over-expression of a bHLH factor triggers excess trichomes differentiation in Arabidopsis, whereas it has no effect in tobacco and tomato (Solanum lycopersicum) [82].
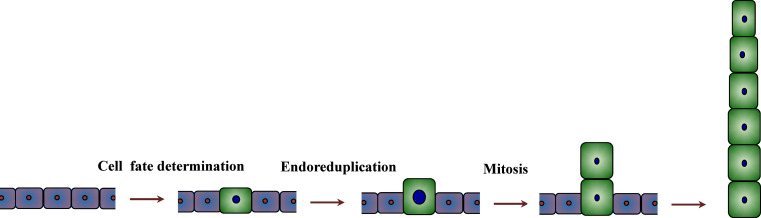
Fig. 2.
Development process of multicellular trichome formation. The formation process of multicellular trichomes is divided into three different stages: cell fate determination, endoreduplication, and mitosis. The epidermal cells destined to become trichomes enlarge first and then divide in line perpendicular to the epidermal surface in the context of continuing cell division
Tomato produces seven types of trichomes, types I–VII, and in tomato woolly mutant type I trichomes increase but other types are not affected, indicating that this type of trichomes develops via distinct regulatory pathways. Woolly (Wo) gene encodes a homeodomain-leucine zipper (HD-Zip) protein and contains a single point mutation [24]. Three alleles were identified at this locus, each of which was with one amino acid replacement, e.g., Arg-to-Leu substitution takes place in Wom, Pro-to-Arg in Wo− and Ile-to-Thr in Womz [24]. As for ETC2 and AtMYC1, the sites of the mutation in these alleles are highly conserved, which possibly change the stability or activity of the proteins [24]. The woolly phenotype was produced in CaMV 35S::Wo transgenic plants and suppression of Wo expression by RNAi decreased the number of type I trichomes [24]. As for MIXTA, the overexpression of Wo in Arabidopsis has no effect on trichome initiation. These molecular data demonstrate that multicellular trichomes in tobacco and tomato and unicellular trichomes in Arabidopsis and cotton are not homologous structures and that their initiations are controlled by different regulatory pathways. For example, in Arabidopsis PROTODERMAL FACTOR2 (PDF2), which has 73 % amino acid sequence identity to Wo, regulates shoot epidermal cell differentiation but not trichome formation [83]. This suggests that during the course of evolution, PDF2 and Wo acquired differential biological functions. In angiosperm phylogeny, Arabidopsis and cotton belong to the Rosids, while tobacco, snapdragon, and tomato belong to the Asterids. Ectopic expression of the bHLH transcription factor LC from maize induced ectopic trichome formation in Arabidopsis, whereas it failed to do so in tobacco and tomato [84]. Phylogenetic analysis indicated that unicellular trichome-related genes GL1 and GaMYB2 fall into one clade, and multicellular trichome-related gene MIXTA and its homologues, into another [81]. Therefore, we infer that regulatory pathways for trichome formation in Rosids and Asterids evolved independently. In another Rosid, poplar (Populus euphratica), overexpression of PtaMYB186, which is correlated with a trichome initiation regulator AtMYB106 in Arabidopsis, increases trichome density [85], and transformation of Arabidopsis GL3 into Brassica napus induces trichome formation on seedling tissues [86]. Taken together, these molecular data demonstrate that species in the Rosids share the common regulatory network, but whether different species in Asterids do is unknown. Different types of multicellular trichomes in Asterids species may be controlled by distinct protein complexes. As stated above, phytohormones modulate unicellular trichome differentiation, but whether the regulatory role of phytohormones in unicellular and multicellular trichome formation is conserved remains unclear. Application of methyl jasmonate could promote the formation of type VI trichomes in tomato [87], and the jasmonic acid-insensitive1 mutant in tomato showed reduced trichome number, indicating JA participates in multicellular trichome formation [88]. In addition, CK and GA also promote trichome formation in tomato, however, different types of trichomes are stimulated by different hormones, without one triggering all types simultaneously. For example, type VI trichome in tomato was specifically activated by JA, and type VII by CK [87]. This is consistent with the notion that in a species with several types of trichomes, different trichome types are controlled by different pathways. Thus, phytohormones play similar promoting roles in the initiation of multicellular trichomes and may share common signaling channels in trichome formation in Rosids and Asterids.
HD-Zip IV transcription factors in trichome formation
HD-Zip proteins are specific to the plant kingdom, with a bZip motif immediately downstream of the HD domain [89]. The former is involved in DNA binding and the latter in protein–protein interactions [90]. Based on their structural characteristics and additional conserved domains, these proteins are classified into four subgroups (HD-Zip I–IV) and members of each subgroup participate in different biological processes [91]. HD-Zip I–III proteins usually play significant roles in abiotic stress response, auxin signaling, embryogenesis, and lateral organ initiation [92]. In this review, we mainly focus on the HD-Zip IV proteins, the majority of which have an epidermis-specific expression pattern [93], and they generally regulate the processes related to epidermal cell differentiation, trichome formation, and anthocyanin synthesis. In maize, outer cell layer4 (OCL4) encodes a HD-Zip IV protein, which not only inhibits trichome patterning in maize but also in Arabidopsis [94]. GaHOX1, characterized by the four conserved domains of the fourth subgroup, is a functional homolog of GL2 in trichome initiation in cotton [76] and shows high amino acid similarities with GbML, which also participates in cotton fiber control [74]. Tomato Wo gene encodes a HD-Zip protein containing a START motif and a SAD domain, which is responsible for the woolly phenotype by activating type I trichome initiation [24]. All alleles at this locus take place in their C terminus, suggesting that this region hides important information for functions and is consistent with the finding that an amino acid replacement in the same region of CD2 results in functional defect [95]. Since there is a close relationship between HD-Zip IV transcriptional factors and epidermal cell differentiation, we consider HD-Zip IV as the common regulator for the formation of unicellular and multicellular trichomes, however, it is not known why the homologous genes are involved in different pathways. During evolution, the four highly conserved domains rarely change, suggesting that diversiform function may lie in the altered part of these genes. As HD-Zip IV members generally act as transcription factors, it is reasonable to infer that they function through binding with other partners and regulating their downstream genes expression, and these putative partners and targets may help us to construct the crosslink in different trichome patterning. SlCycB2, which is similar to a hypothetical B-type cyclin AT5G06270.1 in Arabidopsis, works together with Wo through direct protein–protein interaction in the regulation of the type I trichome formation in tomato [24]. Furthermore, the bZip domain is highly conserved and responsible for the protein-protein interaction, and therefore, we speculate that the interaction between HD-Zip proteins and B type cyclins may be conserved and exist broadly during trichome formation.
Cell cycle regulation in trichome formation
Plant tissue and organ differentiation is closely coordinated with the production of different cell types, involving specific cell fates depending on their position and internal and external signals in plants. Cell fate determination and maintenance mainly lies in the control of the cell cycle, and these processes are usually accompanied by the transition from mitosis to endoreduplication, as evidenced for trichome formation [96]. The number of trichome branches is also positively correlated with the DNA content [97]; elevated ploidy levels result in more branches, whereas reduced levels in fewer branches. Hence, we believe that the basic mechanism for cell cycle control plays an important role in both trichome initiation and branch formation.
Many regulators have been identified for trichome patterning, but whether these genes control trichome initiation and development through regulating the cell cycle is not known. It is noteworthy that two members of the regulatory network discussed above, GL3 and TRY, also participate in the regulation of branch formation. gl3 mutants produce trichomes with reduced branches, whereas try mutants have multi-branch trichomes [10, 41]. This suggests that these two trichome-patterning genes possess dual functions of trichome initiation and branching. In our view, there is a direct relationship between patterning genes and the endoreduplication cycle. A putative plant-specific CDK inhibitor, SIM, inhibits the switch from mitosis to endoreduplication [98] and in sim mutants in Arabidopsis multicellular trichomes are formed with fewer leaf trichomes [99]. Also, mitosis has an opposite effect on the trichome-cell number compared with trichome number, i.e., increased mitotic cycle inhibits trichome initiation and activates the formation of multicellular trichome. Grebe [100] inferred that the inhibition of trichome initiation in Arabidopsis with increased mitosis results from the activating complex not reaching the threshold level to promote trichome cell fate. However, since the cells chosen to be trichomes must exit mitosis and enter the endoreduplication cycle, we consider that the enhancement of mitosis in epidermis may interfere with the exit from mitosis and trichome cell fate determination. This is consistent with the observation that the ectopic expression of an endoreduplication activator CCS52A1 triggers some trichome-like structures in gl2gl3 mutants [101], and the loss-of-function mutations in CCS52A1 enhance the phenotype of sim mutants [102]. Since cell-cycle control acts as downstream targets of the regulatory complex of trichome cell fate and morphogenesis we speculate that this complex induces trichome differentiation by directly or indirectly controlling the expression of some cell cycle-related factors. Based on ChIP experiments, it was indicated that SIM and RBR1 may be the direct targets of two trichome patterning genes GL1 and GL3 through DNA–protein interaction [44] and function-defect mutation in RBR with increased endoreduplication cycles resulted in the formation of multi-branch trichomes [103]. These data provide a direct link between the MYB/bHLH/WD-repeat complex and cell cycle in trichome formation.
Plant cell morphogenesis is determined by the precisely regulated expansion of the cell wall and direction of cell growth, and depends mainly on the cytoskeleton. In Arabidopsis, trichomes are characterized by special cell shapes with three branches, and the incipient trichomes successively undergo four endoreduplication cycles. Trichome branching is coordinated by genes classified into two types based on their different regulatory roles, one type participates in controlling the number of endoreduplication cycles and thus the branch number, and the other affects branch number without altering endoreduplication. Recent results suggest that four independent pathways participate in trichome branching, including endoreduplication, microtubules, Golgi/transcription, as well as STI-dependent processes [104]. The genes related with DNA levels, GL3, TRY, SPY, KAKTUS (KAK), POLYCHOME (PYM), and RASTAFARI (RFI), affect trichome branch number through altering DNA levels [105, 106]. Since both GL3 and TRY act as regulators in trichome initiation and branching, it is of interest to determine whether the rest of these genes are involved in the regulation of trichome patterning. KAK encodes a HECT protein that specifically represses endoreduplication in trichomes through the degradation of specific proteins by an ubiquitin system, implying that ubiquitin-mediated protein degradation functions in trichome branching [105]. Endoreduplication also plays an important role in maintaining cell fate, as reduced endoreduplication causes trichomes to lose their identity [101].
Cell-cycle progression is governed by many cyclin-dependent kinases (CDKs) and the activity of CDKs is associated with their binding activators and inhibitors [107]. Cyclins, as essential activators of CDKs, exists extensively in various species and are classified into many distinct types based on their sequence similarities, which regulate the transition between different phases of the cell cycle. For example, B-type cyclins mainly control the transition of G2-to-M, and D-type the G1-to-S transition [9]. Cyclins not only activate CDKs but also contribute to the substrate specificity of the CDK–cyclin complexes [108]. Distinct CDK–cyclin complexes promote the onset of DNA replication by phosphorylating their substrates at the G2-to-M and G1-to-S transition points [108]. Although many cell cycle regulators have been identified in the past two decades, little is known of their roles in trichome differentiation and development. The ectopic expression of CYCLIN B1;2, which encodes a B-type mitotic cyclin controlling the G2-M transition, induces the formation of multicellular trichomes in Arabidopsis [109]. Interestingly, the sim mutants activate the additional expression of CYCLIN B1;2, whereas wild-type trichomes do not, suggesting that the expression of this gene is normally inhibited by SIM [108] (Fig. 1). Also, specific expression of the D-type cyclin CYCD3;1 in Arabidopsis trichomes induced cell divisions, implying D-type cyclins function similar to B-type cyclins in trichome formation [110]. In addition, as cyclin D can be repressed by a paralog of SIM from rice when expressed in yeast, this suggests that CYCD3;1 may be a second target of SIM [111] (Fig. 1). Strikingly, a B-type cyclin SlCycB2 was also characterized in tomato multicellular trichome formation, which has no obvious similarity with CYCLIN B1;2, but a 53 % sequence identity with AT5G06270.1, a hypothetical B-type cyclin in Arabidopsis [24]. This suggests that SlCycB2 shows a similar function to CYCLIN B1;2, but it is completely a novel gene. Formation of multibranch type V-like trichomes in SlCycB2-RNAi transformants and increased trichome numbers in woolly mutants suggests this gene participates in trichome cell-fate determination and subsequent development [24]. However, the function of AT5G06270.1 in Arabidopsis trichome formation remains unclear. Future studies of SlCycB2 and AT5G06270.1, which may be the common factors in unicellular and multicellular trichome formation, may provide us the potential direct link between two different types of trichomes. Based on all these findings, we conclude that in Arabidopsis, mitosis needs to be inhibited and endoreduplication activated during trichome cell fate determination, and that mitosis remains inhibited during further development, leading to unicellular forms. In tomato and other species that produce multicellular trichomes, there may be a similar regulatory model before trichome initiation, but the pro-trichome cells continue several mitotic cycles, resulting in the formation of multicellular forms (Fig. 3).
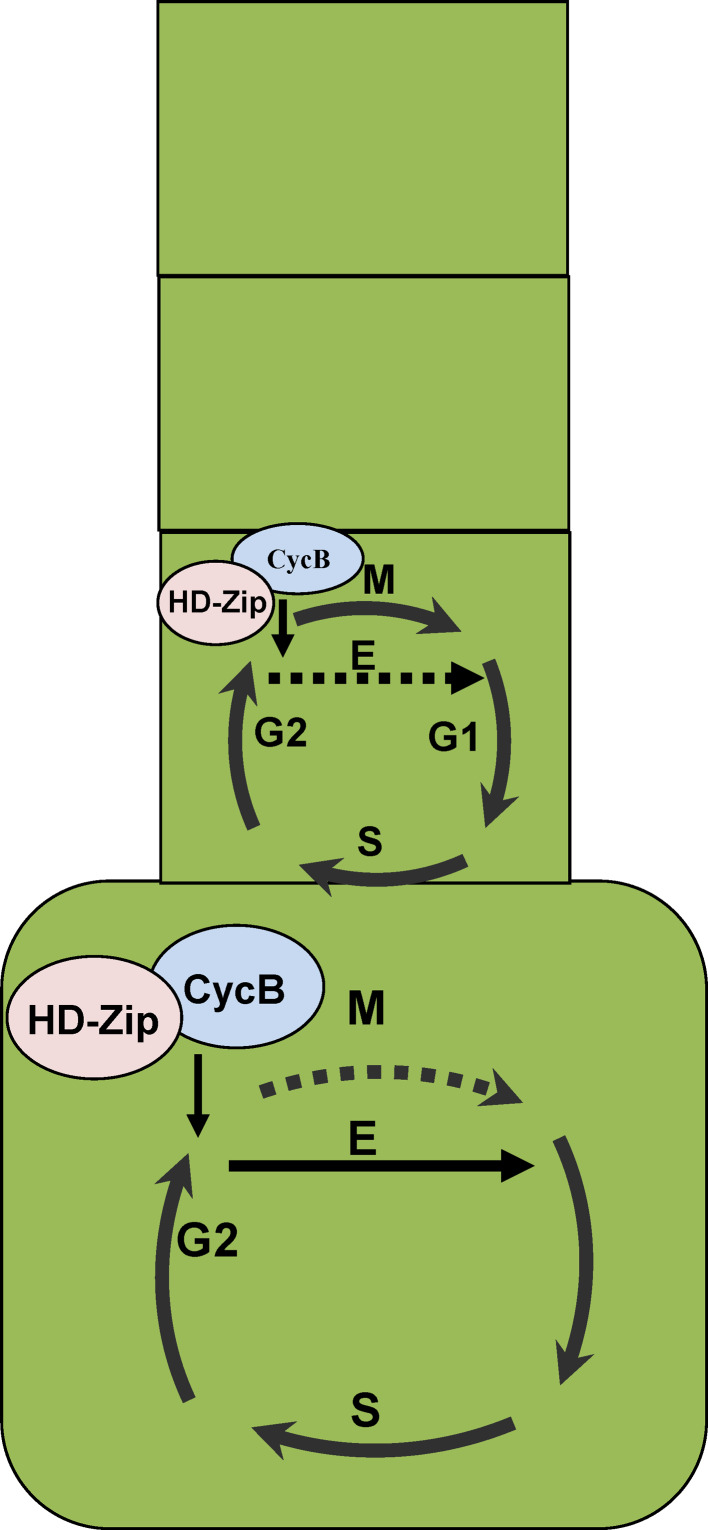
Fig. 3.
Cell-cycle regulation during the process of multicellular trichome formation. In tomato and some other species producing multicellular trichomes, mitosis needs to be repressed and endoreduplication (E) needs to be activated during trichome cell fate determination, and thereafter the protrichome cells will continue several mitosis cycles, resulting in the formation of multicellular forms. Mitosis is inhibited during trichome development, leading to unicellular forms. There may be a similar regulatory model before trichome initiation both in unicellular and multicellular trichome formation, and the dimerization of HD-Zip proteins and B-type cyclins (CycB) must play important roles in these processes
In animals and yeast cells, it was reported that cyclin degradation is essential for cell cycle progression [112]. Apc11p, an E3 ubiquitin ligase, can mediate the ubiquitination of B-type cyclin in vitro [113]. An ubiquitin-conjugating enzyme Ubc4-APC/C can transfer several ubiquitins to cyclin B [114]. Interestingly, the concentration of mitotic cyclins fluctuates throughout the cell cycle [115]. These results demonstrate that cyclins are degraded through the ubiquitin-dependent proteasome pathway. However, little is known about the pathway of cyclin-degradation in plants. KRP1, a cyclin-dependent kinase inhibitor in Arabidopsis, regulates the G1–S transition, and its degradation involves two ubiquitin protein ligases, SCFSKP2 and RKP [116]. Furthermore, some plant cyclins contain a destruction box essential for the cyclin proteolysis by the ubiquitin/proteasome pathway [117]. Thus, it is reasonable to speculate that cyclins and CDKs in trichome formation are also mediated by the same pathway.
Perspectives
The temporal and spatial control of the cell cycle is essential for plant cell differentiation. Distinct cell types arise from precursor cells and become committed to specified cells during differentiation. Plant trichomes are excellent model systems, which can provide valuable information for understanding cell fate determination and maintenance. Although many trichome-patterning genes have been identified and two regulatory models have been proposed, the new findings provide new challenges and require further investigation. Also, the mechanisms by which phytohormone signaling pathways are integrated with the known regulators of trichome formation remain unknown. It would be of interest to ascertain whether the genes responsible for phytohormones synthesis and/or signaling participate in trichome formation and whether phytohormones play similar roles in unicellular and multicellular trichomes. It is clear that both unicellular and multicellular trichomes cannot be formed without the cell cycle control. Therefore, it will be very interesting to reveal whether some cell cycle-related genes, e.g., AT5G06270 and SlCycB2, function as identical activators for different types of trichomes. In addition, as members of HD-Zip IV trigger trichome formation in different species, the sequence characterization and the regulatory information of their four conserved domains will help us determine the roles of different genes. Genetic studies have shown that HD-Zip IV proteins usually function through activating their downstream target genes, and in this case, the ChIP analysis may reveal the unknown targets. Further research is also required to explain whether the protein–protein interaction between HD-Zip IV proteins and CycBs is conserved in the regulation of different types of trichomes.
Acknowledgments
We thank Dr. V·K. Sawhney for critical reading of this manuscript and suggestions. This work was supported by 973 Project Grant 2011CB100600, National Natural Science Foundation Grant 30971997, and China Agricultural Research System Grant CARS-25-A-02.
References
- 1.Werker E. Trichome diversity and development. Adv Bot Res. 2000;31:1–35. [Google Scholar]
- 2.Tingey WM. Potato glandular trichomes: defensive activity against insect attack. ACS Symp Ser. 1991;449:126–135. [Google Scholar]
- 3.Levin DA. The role of trichomes in plant defense. Q Rev Biol. 1973;48:3–15. [Google Scholar]
- 4.Nobel PS (1999) Leaves and fluxes. In: physicochemical and environmental plant physiology, 2nd ed. (Edited by Nobel PS), pp. 293–349. Academic Press, San Diego
- 5.Karabourniotis G, Papadopoulos K, Papamarkou M, Manetas Y. Ultraviolet-B radiation absorbing capacity of leaf hairs. Plant Physiol. 1992;86:414–418. [Google Scholar]
- 6.Wang G, Tian L, Aziz N, Broun P, Dai X, He J, King A, Zhao PX, Dixon RA. Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 2008;148:1254–1266. doi: 10.1104/pp.108.125187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilmillera AL, Schauvinholdb I, Larsonc M, Xub R, Charbonneaua AL, Schmidtb A, Wilkersona C, Lasta RL, Picherskyb E. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang JH, Shi F, Jones AD, M. Marks MA, Howe GA. Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot. 2010;61:1053–1064. doi: 10.1093/jxb/erp370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 10.Hülskamp M, Misera S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis . Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 11.Schnittger A, Hülskamp M. Trichome morphogenesis: a cell-cycle perspective. Philos Trans R Soc Lond Ser B. 2002;357:823–826. doi: 10.1098/rstb.2002.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover BJ, Martin C. Specification of epidermal cell morphology. Adv Bot Res. 2000;31:193–217. [Google Scholar]
- 13.Marks MD, Feldmann KA. Trichome development in Arabidopsis thaliana. I. TDNA tagging of the GLABROUS1 gene. Plant Cell. 1989;1:1043–1050. doi: 10.1105/tpc.1.11.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 15.Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development. 2008;135:1991–1999. doi: 10.1242/dev.016873. [DOI] [PubMed] [Google Scholar]
- 16.Walker AR, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1 . Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis . Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Wang J, Yu N, Li C, Luo B, Gou J, Wang L, Chen X. Control of plant trichome development by a cotton-fiber MYB gene. Plant Cell. 2004;16:2323–2334. doi: 10.1105/tpc.104.024844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payne T, Clement J, Arnold D, Lloyd A. Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum . Development. 1999;126:671–682. doi: 10.1242/dev.126.4.671. [DOI] [PubMed] [Google Scholar]
- 21.Glover BJ, Bunnewell S, Martin C. Convergent evolution within the genus Solanum: the specialised anther cone develops through alternative pathways. Gene. 2004;331:1–7. doi: 10.1016/j.gene.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- 23.Schnittgera A, Folkersa U, Schwaba B, Jürgensa G, Hülskampa M. Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis . Plant Cell. 1999;11:1105–1116. doi: 10.1105/tpc.11.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C, Li H, Zhang J, Luo Z, Gong P, Zhang C, Li J, Wang T, Zhang Y, Lu Y, Ye Z. A regulatory gene induces trichome formation and embryo lethality in tomato. Proc Natl Acad Sci USA. 2011;108:11836–11841. doi: 10.1073/pnas.1100532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schellmann S, Hülskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Dev Biol. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- 26.Pesch M, Hülskamp M. One, two, three… models for trichome patterning in Arabidopsis? Curr Opin Plant Biol. 2009;12:587–592. doi: 10.1016/j.pbi.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis . Annu Rev Plant Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- 28.Pesch M, Hülskamp M. Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Current Opin Genet Dev. 2004;14:422–427. doi: 10.1016/j.gde.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 30.Kang YH, Kirik V, Hülskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell. 2009;21:1080–1094. doi: 10.1105/tpc.108.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E. Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 2007;145:1–11. doi: 10.1104/pp.107.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, Okada K, Wada T. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rerie WG, Feldmann KA, Marks MD. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis . Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 34.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC . Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 36.Kirik V, Simon M, Hülskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis . Dev Biol. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 37.Kirik V, Simon M, Wester K, Schiefelbein J, Hülskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis . Plant Mol Biol. 2004;55:389–398. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen JG. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis . BMC Plant Biol. 2008;8:81. doi: 10.1186/1471-2229-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development. 2008;135:1335–1345. doi: 10.1242/dev.017947. [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Kwak SH, Zeng Q, Ellis BE, Chen X, Schiefelbein J, Chen J. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis . Development. 2007;134:3873–3882. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- 41.Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis . Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Chen J. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2 . Plant Cell Physiol. 2008;49:1792–1804. doi: 10.1093/pcp/pcn159. [DOI] [PubMed] [Google Scholar]
- 43.Balkunde R, Pesch M, Hulskamp M. Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol. 2010;91:299–321. doi: 10.1016/S0070-2153(10)91010-7. [DOI] [PubMed] [Google Scholar]
- 44.Morohashi K, Grotewold E. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 2009;5:e1000396. doi: 10.1371/journal.pgen.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S, Barron C, Schiefelbein J, Chen J. Distinct relationships between GLABRA2 and single repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis . New Phytol. 2010;185:387–400. doi: 10.1111/j.1469-8137.2009.03067.x. [DOI] [PubMed] [Google Scholar]
- 46.Yu N, Cai W, Wang S, Shan C, Wang L, Chen X. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana . Plant Cell. 2010;22:2322–2335. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larkin LC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD. The roles of the GLABROUS1 and TRANPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell. 1994;6:1065–1076. doi: 10.1105/tpc.6.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- 49.Bouyer D, Geier F, Kragler F, Schnittger A, Pesch M, Wester K, Balkunde R, Timmer J, Fleck C, Hülskamp M. Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 2008;6:e141. doi: 10.1371/journal.pbio.0060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balkunde R, Bouyer D, Hülskamp M. Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis . Development. 2011;138:5039–5048. doi: 10.1242/dev.072454. [DOI] [PubMed] [Google Scholar]
- 51.Pesch M, Hülskamp M. Role of TRIPTYCHON in trichome patterning in Arabidopsis . BMC Plant Biol. 2011;11:130. doi: 10.1186/1471-2229-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gan L, Xia K, Chen J, Wang S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis . BMC Plant Biol. 2011;11:176. doi: 10.1186/1471-2229-11-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing S, Miao J, Li S, Qin G, Tang S, Li H, Gu H, Qu L. Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in Arabidopsis . Cell Res. 2010;20:688–700. doi: 10.1038/cr.2010.54. [DOI] [PubMed] [Google Scholar]
- 54.Symonds VV, Hatlestad G, Lloyd A. Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genet. 2011;7:e1002069. doi: 10.1371/journal.pgen.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilscher J, Schlötterer C, Hauser MT. A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr Biol. 2009;19:1747–1751. doi: 10.1016/j.cub.2009.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus . New Phytol. 2011;191:251–263. doi: 10.1111/j.1469-8137.2011.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kryvych S, Nikiforova V, Herzog M, Perazza D, Fisahn J. Gene expression profiling of the different stages of Arabidopsis thaliana trichome development on the single cell level. Plant Physiol Bioch. 2008;46:160–173. doi: 10.1016/j.plaphy.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Lieckfeldt E, Simon-Rosin U, Kose F, Zoeller D, Schliep M, Fisahn J. Gene expression profiling of single epidermal, basal and trichome cells of Arabidopsis thaliana . J Plant Physiol. 2008;165:1530–1544. doi: 10.1016/j.jplph.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 59.Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana . Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- 60.Chien JC, Sussex IM. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;111:1321–1328. doi: 10.1104/pp.111.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perazza D, Vachon G, Herzog M. Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis . Plant Physiol. 1998;117:375–383. doi: 10.1104/pp.117.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobsen SE, Olszewski NE. Mutations in the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Traw MB, Bergelson J. Interactive effects of jasmonic acid, salicylic acid and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 2003;133:1367–1375. doi: 10.1104/pp.103.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gan Y, Kumimoto R, Chang L, Ratcliffe O, Hao Y, Broun P. GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis . Plant Cell. 2006;18:1383–1395. doi: 10.1105/tpc.106.041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan Y, Liu C, Yu H, Broun P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development. 2007;134:2073–2081. doi: 10.1242/dev.005017. [DOI] [PubMed] [Google Scholar]
- 67.Zhou Z, An L, Sun L, Zhu S, Xi W, Broun P, Yu H, Gan Y. Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis . Plant Physiol. 2011;157:673–682. doi: 10.1104/pp.111.180281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana . Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshida Y, Sano R, Wada T, Takabayashi J, Okada K. Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis . Development. 2009;136:1039–1048. doi: 10.1242/dev.030585. [DOI] [PubMed] [Google Scholar]
- 70.Qin Y, Zhu Y. How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol. 2011;14:106–111. doi: 10.1016/j.pbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 71.Loguerico LL, Zhang JQ, Wilkins TA. Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.) Mol Gen Genet. 1999;261:660–671. doi: 10.1007/s004380050009. [DOI] [PubMed] [Google Scholar]
- 72.Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 2009;59:52–62. doi: 10.1111/j.1365-313X.2009.03847.x. [DOI] [PubMed] [Google Scholar]
- 73.Walford SA, Wu Y, Llewellyn DJ, Dennis ES. GhMYB25-like: a key factor in early cotton fibre development. Plant J. 2011;65:785–797. doi: 10.1111/j.1365-313X.2010.04464.x. [DOI] [PubMed] [Google Scholar]
- 74.Zhang F, Zuo K, Zhang J, Liu X, Zhang L, Sun X, Tang K. An L1 box binding protein, GbML1, interacts with GbMYB25 to control cotton fibre development. J Exp Bot. 2010;61:3599–3613. doi: 10.1093/jxb/erq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Humphries JA, Walker AR, Timmis JN, Orford SJ. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol Biol. 2005;57:67–81. doi: 10.1007/s11103-004-6768-1. [DOI] [PubMed] [Google Scholar]
- 76.Guan X, Li Q, Shan C, Wang S, Mao Y, Wang L, Chen X. The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2 . Physiol Plantarum. 2008;134:174–182. doi: 10.1111/j.1399-3054.2008.01115.x. [DOI] [PubMed] [Google Scholar]
- 77.Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X (2011) GbPDF1 (Protodermal factor 1) is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol, dx.doi.org/10.1104/pp.111.186742 [DOI] [PMC free article] [PubMed]
- 78.Vermeer J, Peterson RL. Glandular trichomes on the inflorescence of Chrysanthemum morifolium cv. Dramatic (Compositae). 2 Ultrastructure and histochemistry. Can J Bot. 1979;57:714–729. [Google Scholar]
- 79.Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Anthirrhinum majus flowers. Development. 2005;132:359–379. doi: 10.1242/dev.01584. [DOI] [PubMed] [Google Scholar]
- 80.Avila A, Nieto C, Cañas L, Benito MJ, Paz-Ares J. Petunia hybrida genes related to the maize regulatory C1 gene and to the animal myb proto-oncogenes. Plant J. 1993;3:553–562. doi: 10.1046/j.1365-313x.1993.03040553.x. [DOI] [PubMed] [Google Scholar]
- 81.Serna L, Martin C. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11:274–280. doi: 10.1016/j.tplants.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Lloyd AM, Walbot V, Davis RW. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science. 1992;258:1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- 83.Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis . Development. 2003;130:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- 84.Glover BJ, Perez-Rodriguez M, Martin C. Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development. 1998;125:3497–3508. doi: 10.1242/dev.125.17.3497. [DOI] [PubMed] [Google Scholar]
- 85.Plett JM, Wilkins O, Campbell MM, Ralph SG, Regan S. Endogenous overexpression of Populus MYB186 increases trichome density, improves insect pest resistance, and impacts plant growth. Plant J. 2010;64:419–432. doi: 10.1111/j.1365-313X.2010.04343.x. [DOI] [PubMed] [Google Scholar]
- 86.Gruber MY, Wang S, Ethier S, Holowachuk J, Bonham-Smith PC, Soroka J, Lloyd A. “HAIRY CANOLA’’–Arabidopsis GL3 induces a dense covering of trichomes on Brassica napus seedlings. Plant Mel Biol. 2006;60:679–698. doi: 10.1007/s11103-005-5472-0. [DOI] [PubMed] [Google Scholar]
- 87.Maes L, Goossens A. Hormone-mediated promotion of trichome initiation in plants is conserved but utilizes species- and trichome-specific regulatory mechanisms. Plant Signal Behav. 2010;5:205–207. doi: 10.4161/psb.5.2.11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schena M, Davis RW. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA. 1992;89:3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee YH, Chun JY. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol Biol. 1998;37:377–384. doi: 10.1023/a:1006084305012. [DOI] [PubMed] [Google Scholar]
- 91.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 92.Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav. 2009;4:86–88. doi: 10.4161/psb.4.2.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis . Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vernoud V, Laigle G, Rozier F, Meeley RB, Perez P, Rogowsky PM. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009;59:883–894. doi: 10.1111/j.1365-313X.2009.03916.x. [DOI] [PubMed] [Google Scholar]
- 95.Isaacson T, Kosma DK, Matas AJ, Buda GJ, He Y, Yu B, Pravitasari A, Batteas JD, Stark RE, Jenks MA, Rose JKC. Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J. 2009;60:363–377. doi: 10.1111/j.1365-313X.2009.03969.x. [DOI] [PubMed] [Google Scholar]
- 96.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 97.Folkers U, Berger J, Hülskamp M. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124:3779–3786. doi: 10.1242/dev.124.19.3779. [DOI] [PubMed] [Google Scholar]
- 98.Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, Gwin T, Churchman J, Larkin JC. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana . Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- 100.Grebe M. The patterning of epidermal hairs in Arabidopsis-updated. Curr Opin Plant Biol. 2011;15:1–7. doi: 10.1016/j.pbi.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 101.Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A. Endoreplication controls cell fate maintenance. PLoS Genet. 2010;6:e1000996. doi: 10.1371/journal.pgen.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kasili R, Walker JD, Simmons LA, Zhou J, De Veylder L, Larkin JC. SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics. 2010;185:257–268. doi: 10.1534/genetics.109.113274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 2006;140:67–80. doi: 10.1104/pp.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hülskamp M. Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- 105.Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003;35:729–742. doi: 10.1046/j.1365-313x.2003.01844.x. [DOI] [PubMed] [Google Scholar]
- 106.Perazza D, Herzog M, Hülskamp M, Brown S, Dorne AM, Bonneville JM. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics. 1999;152:461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Inze D, Veylder LD. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 108.Peeper DS, Parker LL, Ewen ME, Toebes M, Hall FL, Xu M, Zantema A, van der Eb AJ, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1955. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schnittger A, Schöbinger U, Stierhof YD, Hülskamp M. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol. 2002;12:415–420. doi: 10.1016/s0960-9822(02)00693-0. [DOI] [PubMed] [Google Scholar]
- 110.Schnittger A, Schöbinger U, Bouyer D, Weinl C, Stierhof YD, Hülskamp M. Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc Natl Acad Sci USA. 2002;99:6410–6415. doi: 10.1073/pnas.092657299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, Magyar Z, Hatzfeld Y, Schueren EVD, Beemster GTS, Frankard V, Larkin JC, Inzé D, Veylder LD. Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem. 2007;282:25588–25596. doi: 10.1074/jbc.M703326200. [DOI] [PubMed] [Google Scholar]
- 112.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 113.Leverson JD, Joazeiro CAP, Page AM, Huang H, Hieter P, Hunter T. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 115.Richter-Ruoff B, Wolf DH. Proteasome and cell cycle: evidence for a regulatory role of the protease on mitotic cyclins in yeast. FEBS Lett. 1993;336:34–36. doi: 10.1016/0014-5793(93)81603-w. [DOI] [PubMed] [Google Scholar]
- 116.Ren H, Santner A, del Pozo JC, Murray JAH, Estelle M. Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J. 2008;53:705–716. doi: 10.1111/j.1365-313X.2007.03370.x. [DOI] [PubMed] [Google Scholar]
- 117.Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, de Pamphilis CW, Ma H. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 2004;135:1084–1099. doi: 10.1104/pp.104.040436. [DOI] [PMC free article] [PubMed] [Google Scholar]