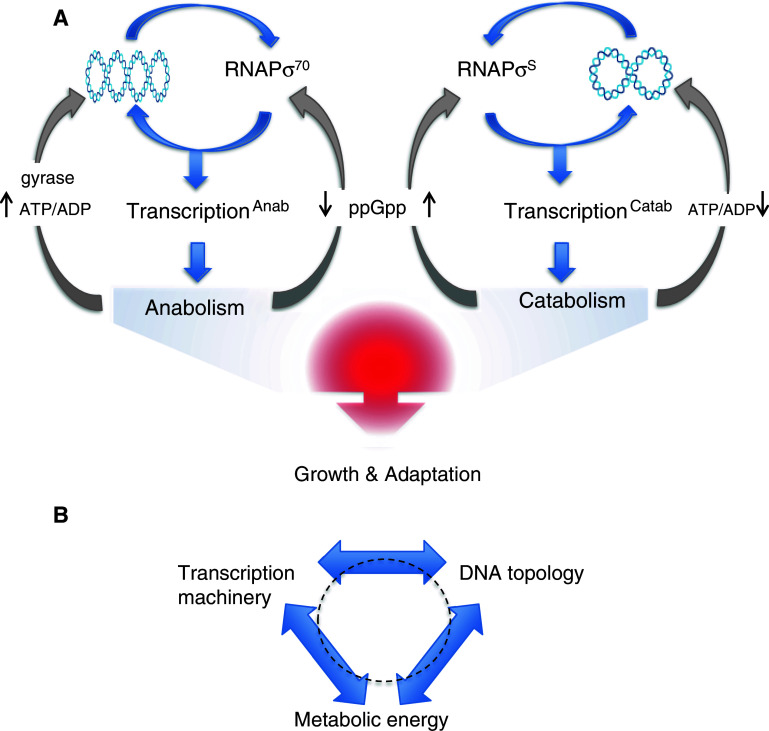
Fig. 3.
Structural coupling between the composition of RNA polymerase holoenzyme, DNA topology and metabolism in E. coli. a The chromosomal DNA is depicted as an interwound “plectonemic” coil. Preference of the vegetative and stationary phase RNA polymerase holoenzymes (RNAPσ70 and RNAPσS, respectively) for correspondingly high or low DNA superhelical density (indicated by changing number of crossings in the DNA plectoneme) specifies either anabolic or catabolic gene expression. The coordination of transcriptional response is mediated by structural coupling between RNAP composition and the DNA topology, whereas both respond to changes of metabolism: increased ATP/ADP ratio supports gyrase activity and high negative superhelicity, whereas increased concentration of the “stringent response” regulator ppGpp supports RNAPσS formation and so the preference for relaxed DNA templates [39]. b Three basic components underlying the irreducible organisational complexity of any living cell. Note that the organisation is essentially circular with all three basic components standing in relationship to reciprocal determination