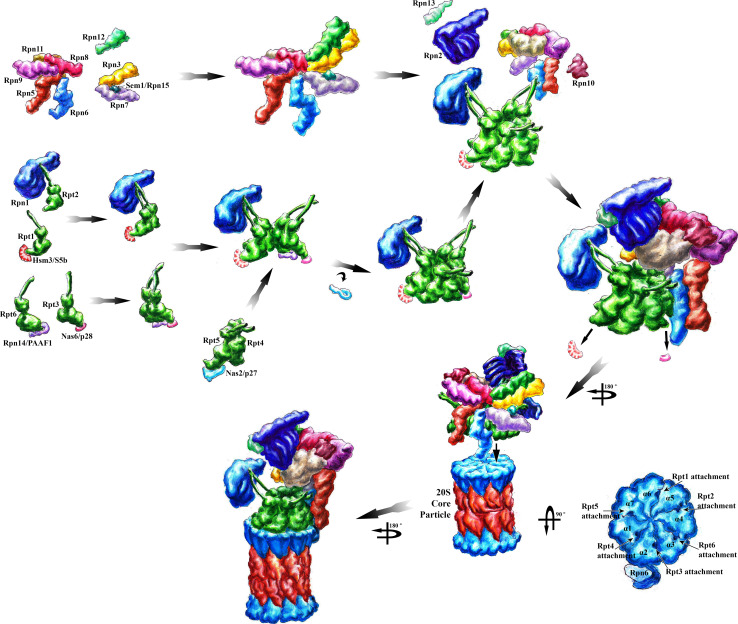
Fig. 3.
The model of CP-independent RP assembly is depicted. In the CP-independent assembly pathway, four RP-dedicated chaperones join RP base modules: Hsm3/S5b binds to the ATPase subunits Rpt1 and Rpt2, Nas2/p27 to Rpt5, Nas6/p28 to Rpt3 and Rpn14/PAAF1 to Rpt6, respectively, by forming three modules or precursors, namely, the Nas6/p28-Rpt3–Rpt6-Rpn14/PPAF1 module, the Nas2/p27-Rpt5–Rpt4 module and the Hsm3/S5b-Rpt1–Rpt2-Rpn2 module. The chaperones are dislocated with the attachment to the CP. The RP lid is assembled from two modules, the Rpn5, 6, 8, 9 and 11 module and the Rpn3, 7 and Sem1 module. The incorporation of Rpn12 completes RP lid assembly. RP base and lid associate with Rpn10 and bind to the CP. Rpn6 also stabilizes the RP–CP interaction. In the CP-dependent assembly pathway, the RP base modules successively dock onto the CP template. The lid modules are assembled on top of the CP–RP base. In the CP-dependent model, the chaperones are most likely released, when the RP base is docking on the CP template to avoid chaperone collisions with the CP–RP base interface. Aberrant RP–CP assemblies are preferentially recognized by Ecm29, which may contact the RP and CP. The structure of Ecm29-bound RP–CP assemblies is unknown. Structures of the proteasomal subunits and proteasomal chaperones were deduced from single particle electron cryo-microscopy and crystal structures deposited in the PDB database. See main text for details