Fig. 2.
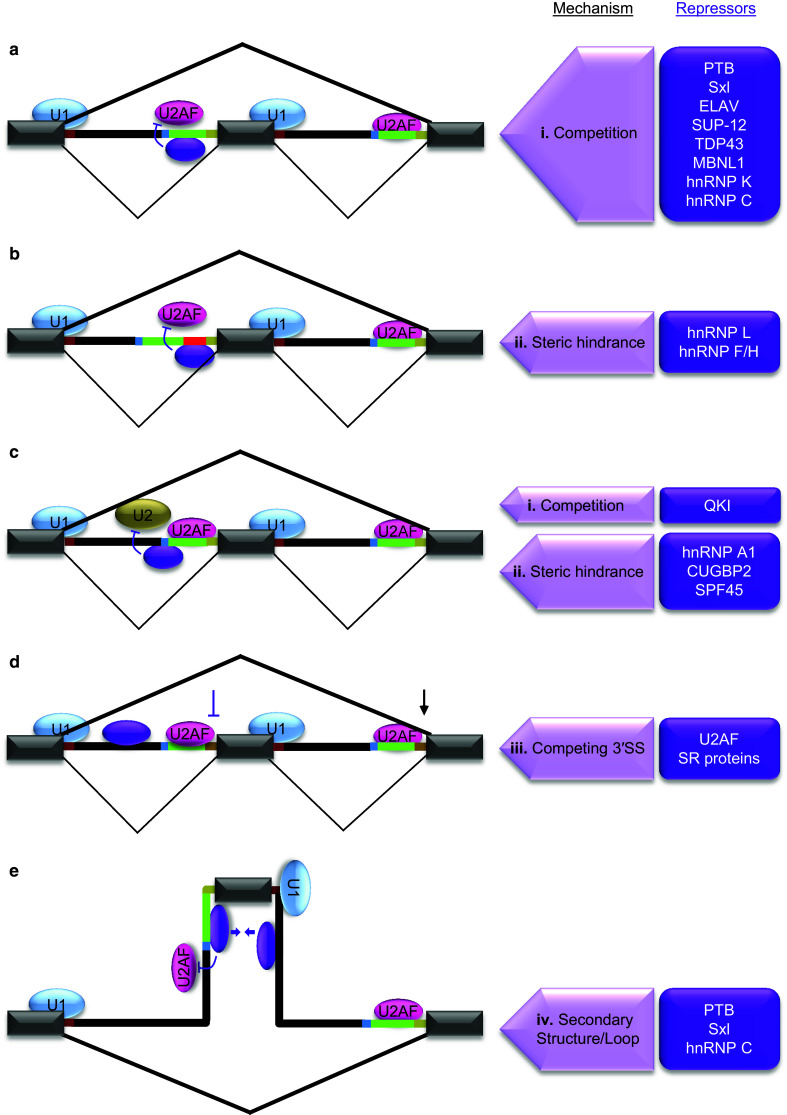
Mechanisms of splicing repression at 3′SS. The regulated exon shown in the middle is flanked by introns and exons. Splicing patterns in bold lines indicate the outcome of regulation. Purple oval represents one of the repressors listed in purple boxes on the right side that are involved in the regulation through respective mechanisms indicated on the left side. a Splicing repression through cis-acting elements within Py. The location of a major class of silencers within 3′SS overlaps with Py. These elements recruit trans-acting factors to inhibit U2AF binding to Py through competition (i). b Splicing repression through cis-acting element insertions between Py and 3′AG. A group of 3′SS silencers is uniquely positioned between Py and 3′AG (red line in intron). Such elements also inhibit U2AF mainly through steric hindrance (ii) caused by specific binding of repressors. c Splicing repression through cis-acting elements near BP. Some 3′SS silencers are located near or in overlap with BP. The binding of trans-acting factors to these elements results in inhibition of U2 snRNP interaction with BP either through competition (i) or steric hindrance (ii). d Inhibition of weakened 3′SS by competition of downstream stronger 3′SS. The weakening of upstream 3′SS by intronic binding of U2AF (purple oval) results in inhibition by competing 3′SS (iii). e Splicing repression through ‘RNA secondary structures or loops’. In some cases Py binding trans-acting alternative splicing factors interact cooperatively with another RNA-binding protein bound to downstream intron (purple ovals and arrows). This interaction results in looping out (iv) of the regulated exon and interferes with U1 and U2AF binding to its splice sites leading to inhibition of exon definition. RNA secondary structures also trap 3′SS motifs in a similar way but without proteins to repress splicing
