Fig. 3.
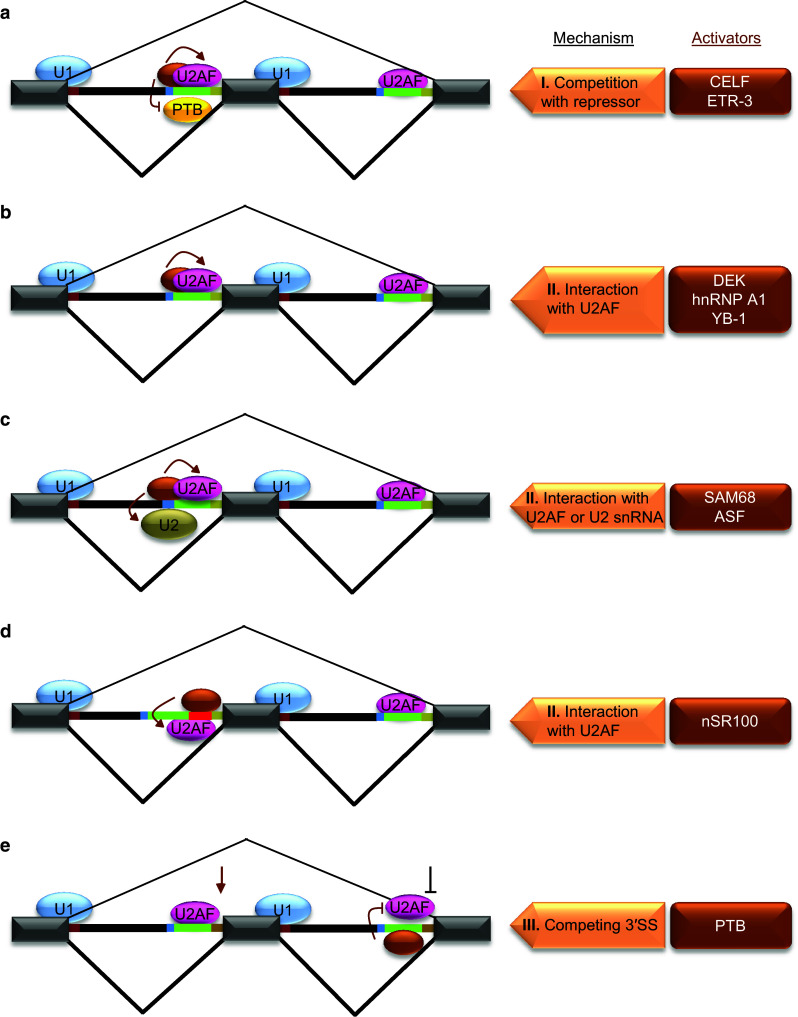
Mechanisms of splicing activation at 3′SS. Individual activators are shown as brown ovals and they are listed on the right side in brown boxes along with respective mechanisms for the regulation shown on the left side. a Splicing activation through inhibition of a repressor by enhancer motifs in Py. The binding of a trans-acting factor to an enhancer motif within Py results in competition with a repressor (yellow oval/PTB) binding (I) and facilitates U2AF binding to Py to promote 3′SS usage. b–d Splicing activation through cis-acting elements located at different positions within 3′SS. The locations of these elements are within Py (b), near BP (c) or between Py and 3′AG (red line) (d). A common feature of 3′SS regulation by these elements is their recognition by respective trans-acting factors for interaction with U2AF or U2 snRNA (II) to facilitate their RNA-binding leading to activation of splicing. e Activation of 3′SS by repression of downstream 3′SS. The repression of a downstream 3′SS through binding of PTB to Py inhibiting U2AF results in activation of a competing 3′SS (III)
