Fig. 4.
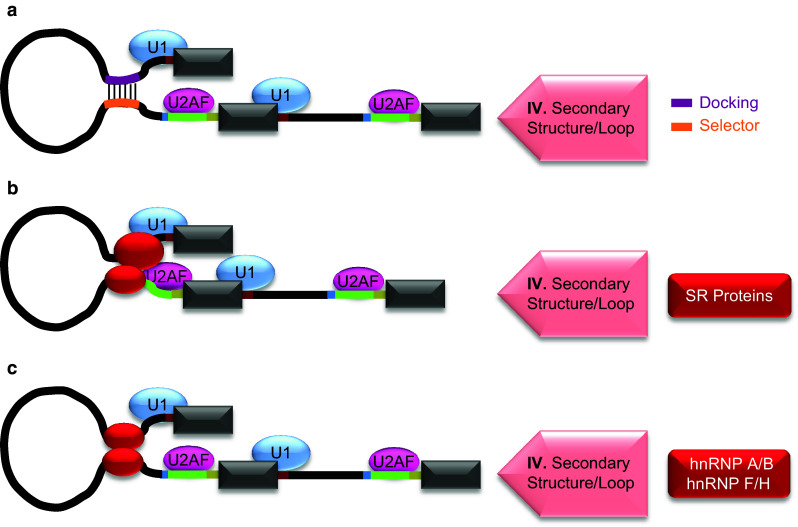
Mechanisms of weaker 3′SS activation through RNA secondary structures/loops. a Base-pairing interaction between intronic RNA motifs. The activation of a 3′SS through looping of pre-mRNA can also result from base-pairing of an intronic motif (purple line) with a second motif located near regulated splice site (orange line). These motifs are referred to as docking or selector sites as indicated on the right side. The formation of such secondary structures or loops (IV) promotes 3′SS usage. b Interaction between a complex of splicing factors and 3′SS spliceosome components. In this mechanism a protein complex (red oval) binds to an intron upstream of regulated exon and interacts with spliceosome proteins of 3′SS to promote its usage. SR proteins shown in red box on the right side form such known complexes. c Cooperative interaction of a splicing factor. This mechanism of pre-mRNA looping involves the binding of a trans-acting splicing factor (red oval) near intron ends. Examples of such factors are shown in red box on the right side. The cooperative interaction between the factors bound near intron ends creates a loop of spanning intronic sequence that brings its splice sites along with their spliceosomal components in close proximity to activate 3′SS
