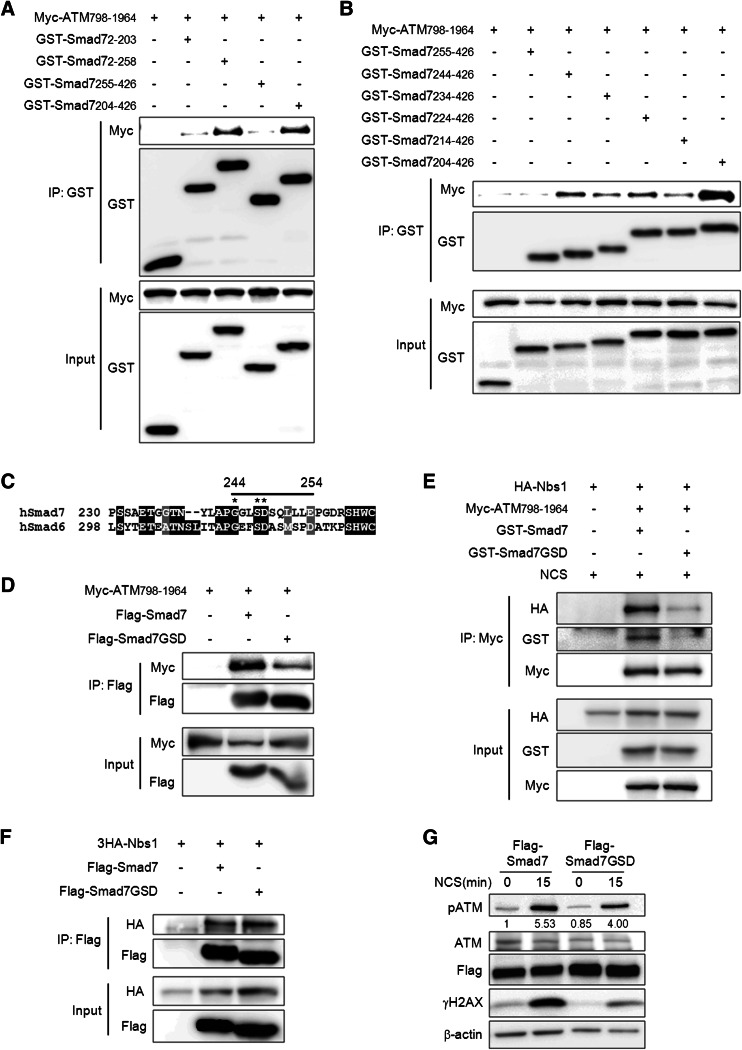
Fig. 5.
Identification of the binding sites that mediate the interaction of ATM and Smad7. a, b To identify the region of Smad7 responsible for its interaction with ATM 798-1964, Smad7 fragments described in supplemental material Fig. S5 were pulled-down using Glutathione Sepharose 4b beads, and a Myc-specific antibody was used to detect the interacting ATM. c Alignment of hSmad7 and hSmad6. The straight black line indicates amino acids 244–254 of Smad7. *indicate sites that were mutated. d To identify the sites that are important for the interaction of Smad7 and ATM-798-1964, we mutated three sites (G244A, S247A, and D248A) on Smad7 and examined the interactions of the mutated proteins with ATM 798-196. Whole cell lysates from the wild type or mutant (Smad7-GSD) Smad7 were immunoprecipitated using an anti-Flag antibody, and western blotting was performed using an anti-Myc antibody to detect the interacting ATM. e Cells were transfected with Myc-ATM-798-1964, 3HA-Nbs1 and GST-Smad7 or GST-Smad7-GSD mutant. The cells were treated with 200 ng/ml NCS to induce DSB. Whole cell lysates were immunoprecipitated using anti-Myc antibody and western blotting was performed using indicated antibody. f To identify whether Smad7-GSD mutant also affect the interactions with Nbs1, whole cell lysates from the wild type or Smad7-GSD mutant was immunoprecipitated using an anti-Flag antibody, and western blotting was performed using anti-HA antibody to detect the interacting Nbs1. g Wild or mutant (Smad7-GSD) Smad7 construct was transiently transfected into A549 cells. Cells were treated with 200 ng/ml NCS for 15 min and the expression of phospho-ATM or γH2AX was examined. Equal protein loading was confirmed by β-actin detection. Ratio pATM/ATM was calculated by densitometric analysis