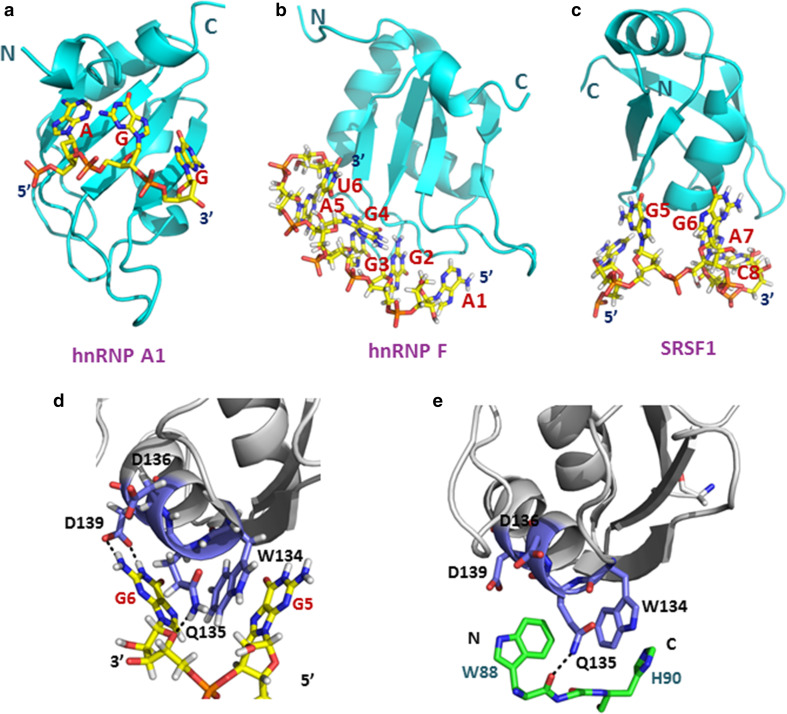
Fig. 6.
Three subclasses of RRMs use different mechanisms for RNA binding. a Structure of hnRNP A1 RRM1 bound to single-stranded telomeric DNA (PDB accession code: 2up1). b Structure of hnRNP F qRRM2 bound to 5′-AGGGAU-3′ RNA (PDB accession code: 2KG0). c Structure of the SRSF1 pseudo-RRM bound to 5′-AGGAC-3′ RNA (PDB accession code: 2M8D). DNA and RNAs are shown in stick representations with carbons in yellow, nitrogens in blue, oxygens in red, and phosphorous in orange. RRM motifs are shown as cartoon models in cyan. d Close-up view of interactions of the SRSF1 pseudo-RRM bound to the GG dinucleotide of the 5′-UGAAGGAC-3′ RNA. e Close-up view of the structure of the SRSF1 pseudo-RRM bound to the Trp-Gly-His tripeptide of SRPK1 (PDB accession code: 3BEG). The side-chains of W88 and H90 occupy the same sites as G6 and G5, respectively. Both the G6 base and the side-chains of W88 and H90 could interact with SRSF1 via hydrogen bond formation. The RRMs are shown in gray with residues interacting with the RNA or peptide presented as stick models in dark blue. The Trp-Gly-His tripeptide is presented as stick model in green, with hydrogen bonds are indicated as black dashed lines