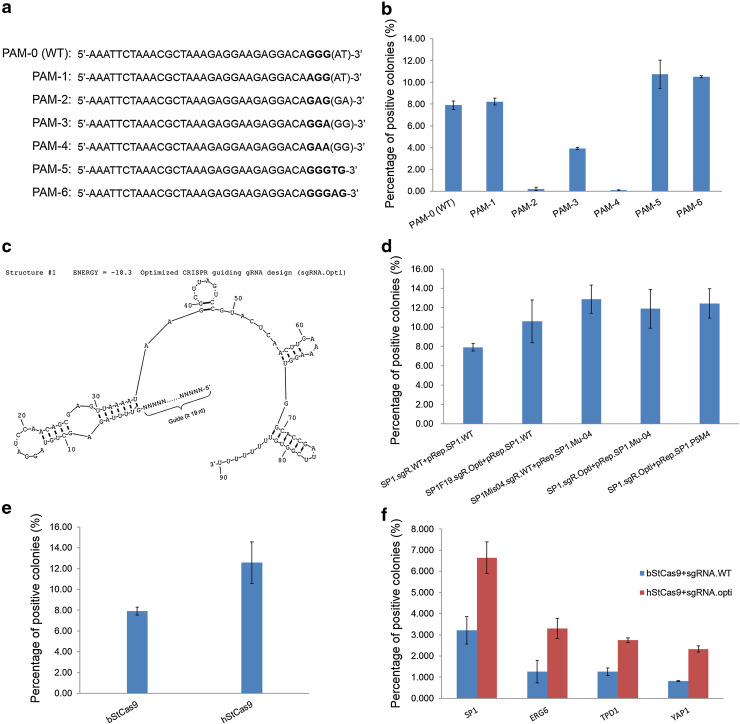
Fig. 4.
The optimized StCas9 system functions in yeast. a Different PAM patterns designed in reporter vectors for SP1 protospacer targeting. b Comparison of different PAM patterns for the StCas9 activity. c Secondary structure of the optimized CRISPR guiding RNA (sgRNA.Opti) design. The structure was drawn by RNAstructure Version 4.6 not accounting for the influence of the guide sequence. d Comparison of wild type and optimized sgRNA designs for SP1 protospacer targeting. e Comparison of the bacterial origin bStCas9 and codon-optimized hStCas9 in yeast assay. f Functional analysis of the StCas9 system on different chromosome integrated targets