Fig. 2.
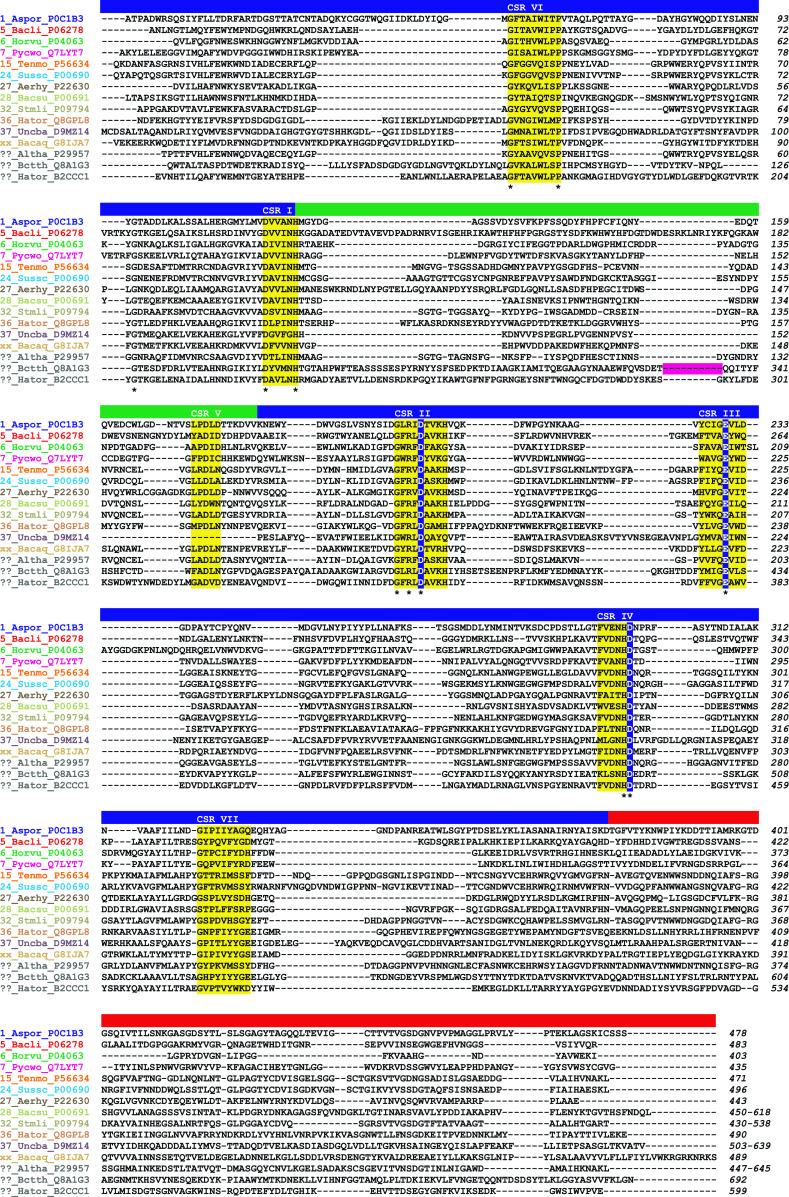
Amino acid sequence alignment of GH13 α-amylases representing the individual α-amylase subfamilies. The aligned sequences span the typical GH13 α-amylase’s domain arrangement consisting of catalytic (β/α)8-barrel with domain B (inserted between the strand β3 and helix α3) and domain C (succeeding the TIM-barrel). The name of an enzyme is composed of the GH13 subfamily number (“xx” for the recently described α-amylase from Bacillus aquimaris and “??” for α-amylases from A. haloplanktis, Bacteroides thetaiotaomicron, and Halothermothrix orenii AmyB that currently are still not assigned any GH13 subfamily in CAZy) followed by the abbreviation of the source (organism) and the UniProt accession number. Note there are two α-amylases from Halothermothrix orenii, the AmyA assigned to subfamily GH13_36 (UniProt Q8GPL8) and the AmyB not assigned as yet to any GH13 subfamily (UniProt B2CCC1). The organisms are abbreviated as follows: Aspor, Aspergillus oryzae; Bacli, Bacillus licheniformis; Horvu, Hordeum vulgare; Pycwo, Pyrococcus woesei; Tenmo, Tenebrio molitor; Sussc, Sus scrofa (pancreas); Aerhy, Aeromonas hydrophila; Bacsu, Bacillus subtilis; Stmli, Streptomyces limosus; Hator, Halothermothrix orenii; Uncba, uncultured bacterium; Bacaq, Bacillus aquimaris; Altha, A. haloplanktis; and Bctth, Bacteroides thetaiotaomicron. The seven CSRs typical for the family GH13 [46] and the catalytic triad are highlighted in yellow and blue, respectively. The residues conserved invariantly are marked by an asterisk below the alignment. The individual α-amylase family GH13 domains are indicated as a colored lane above the alignment: blue catalytic domain A, green domain B, red C-terminal domain C. The positions corresponding to the deleted SBD of the family CBM58 in ??_Bctth_Q8A1G3 (SusG α-amylase) are signified by a magenta lane. All sequences were retrieved from the UniProt knowledge database [251]. The sequence alignments were done using the program Clustal-W2 [252] and then manually tuned in order to maximize sequence similarities
