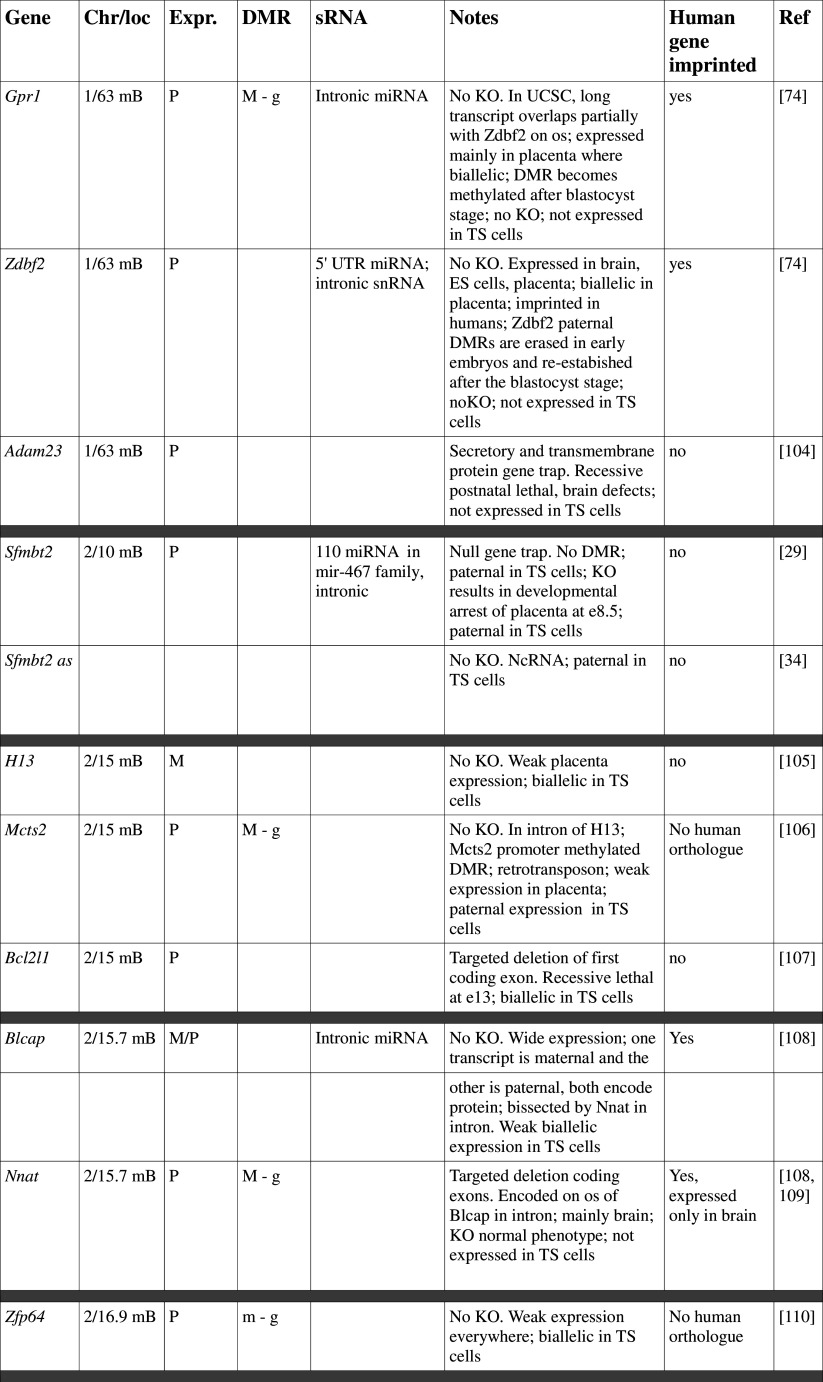
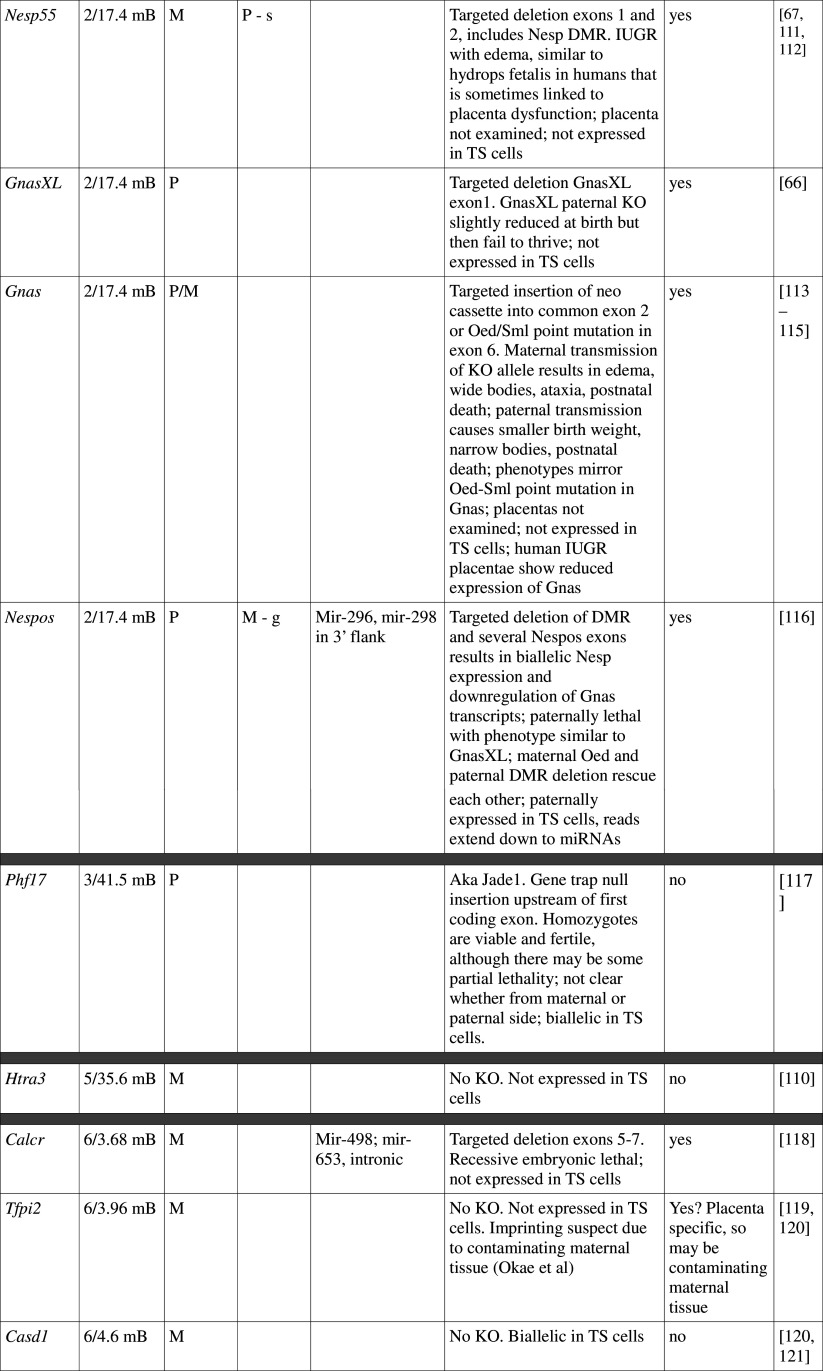
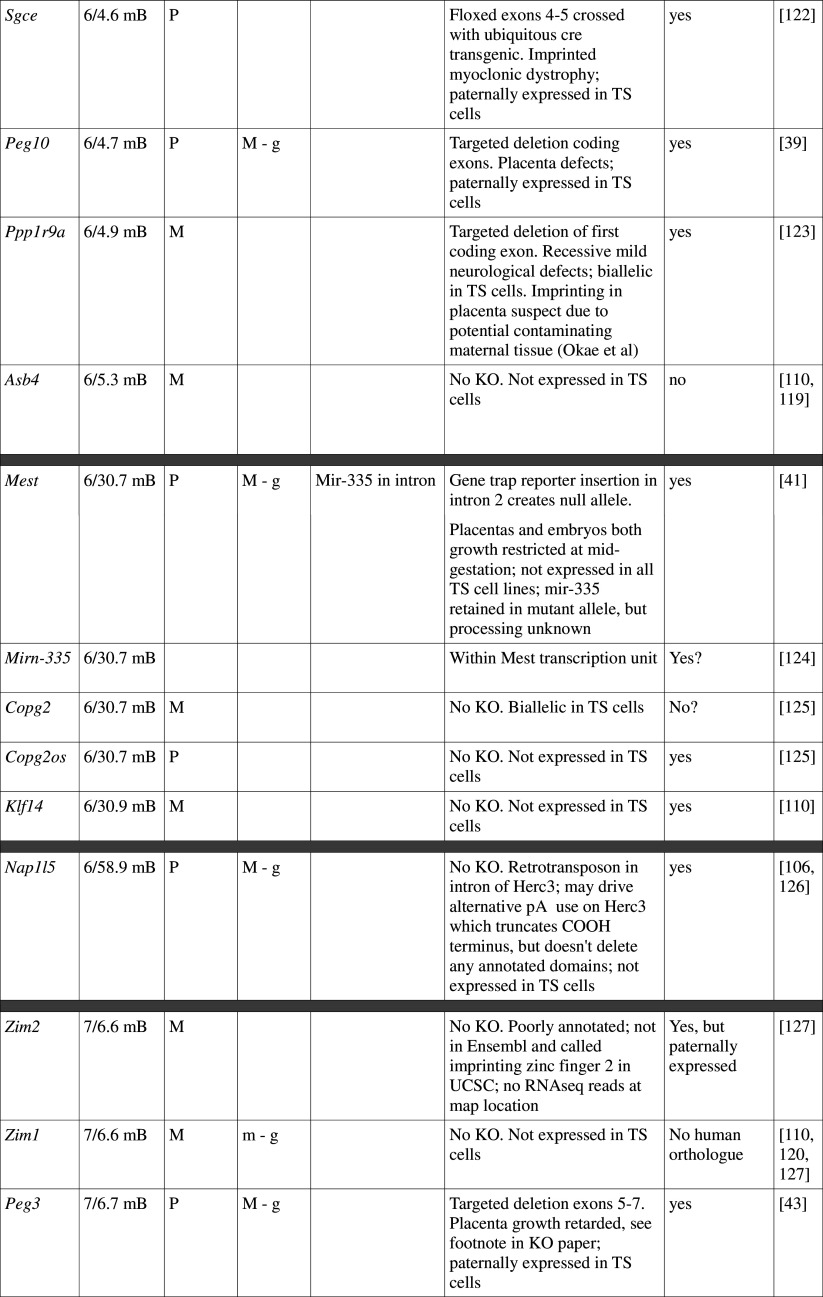
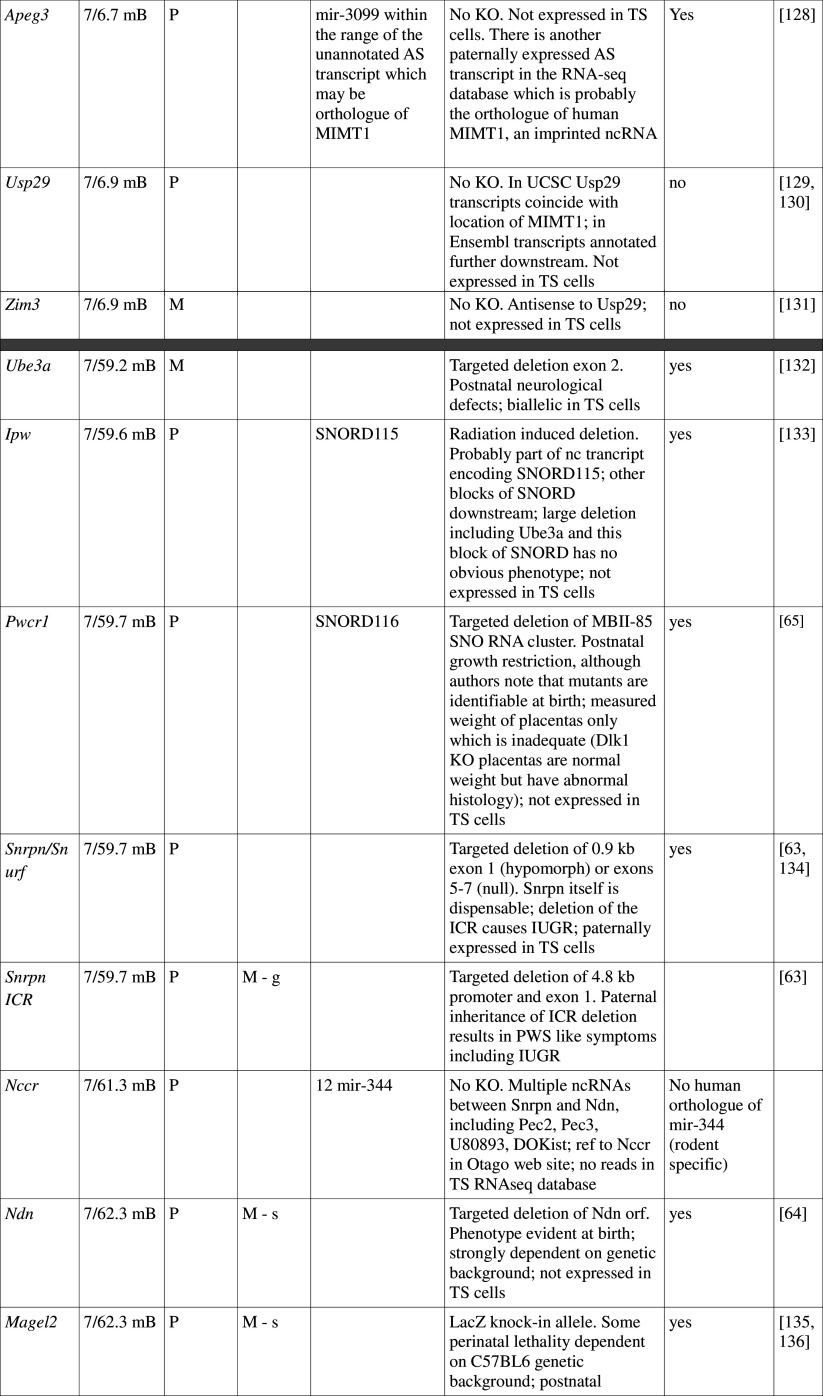
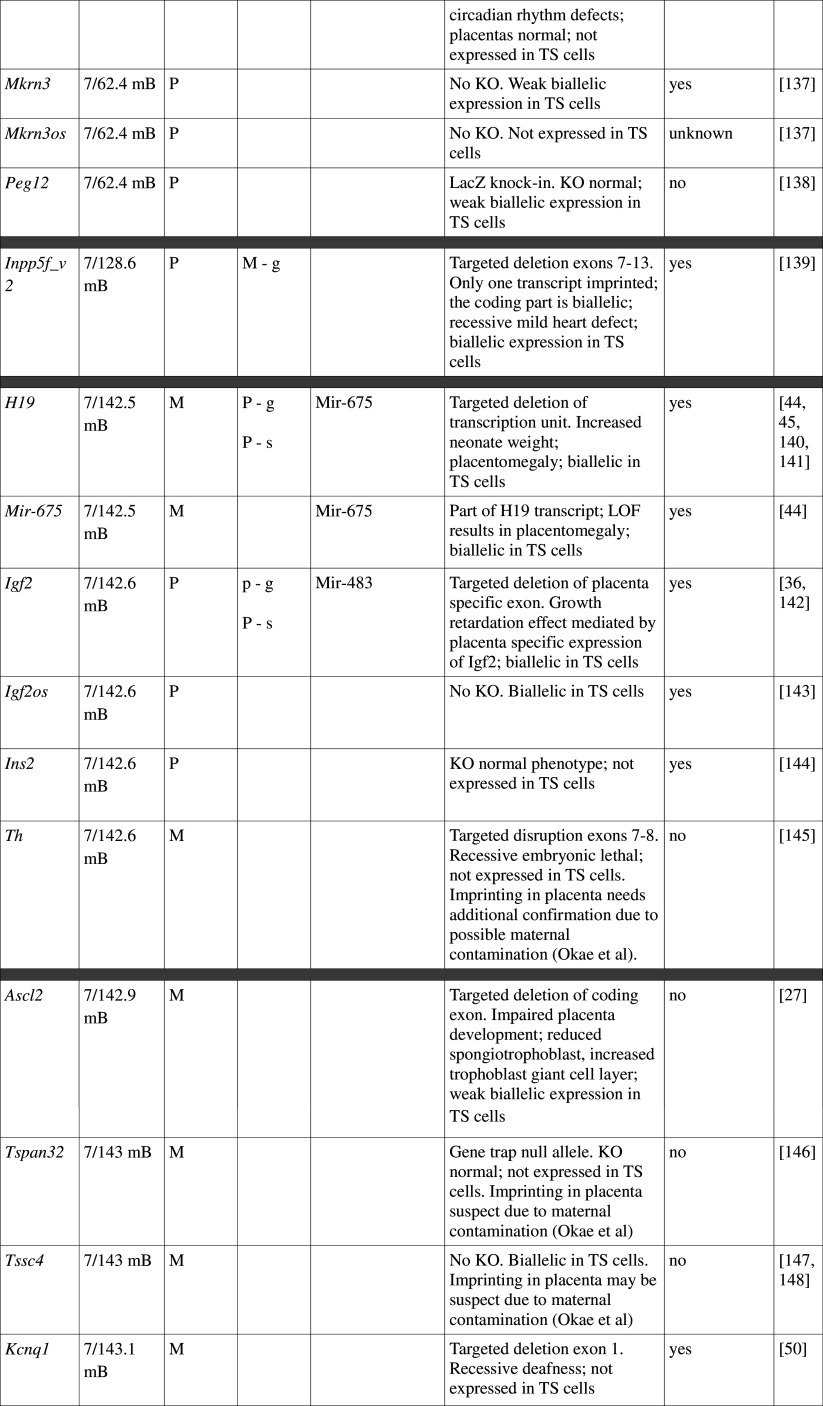
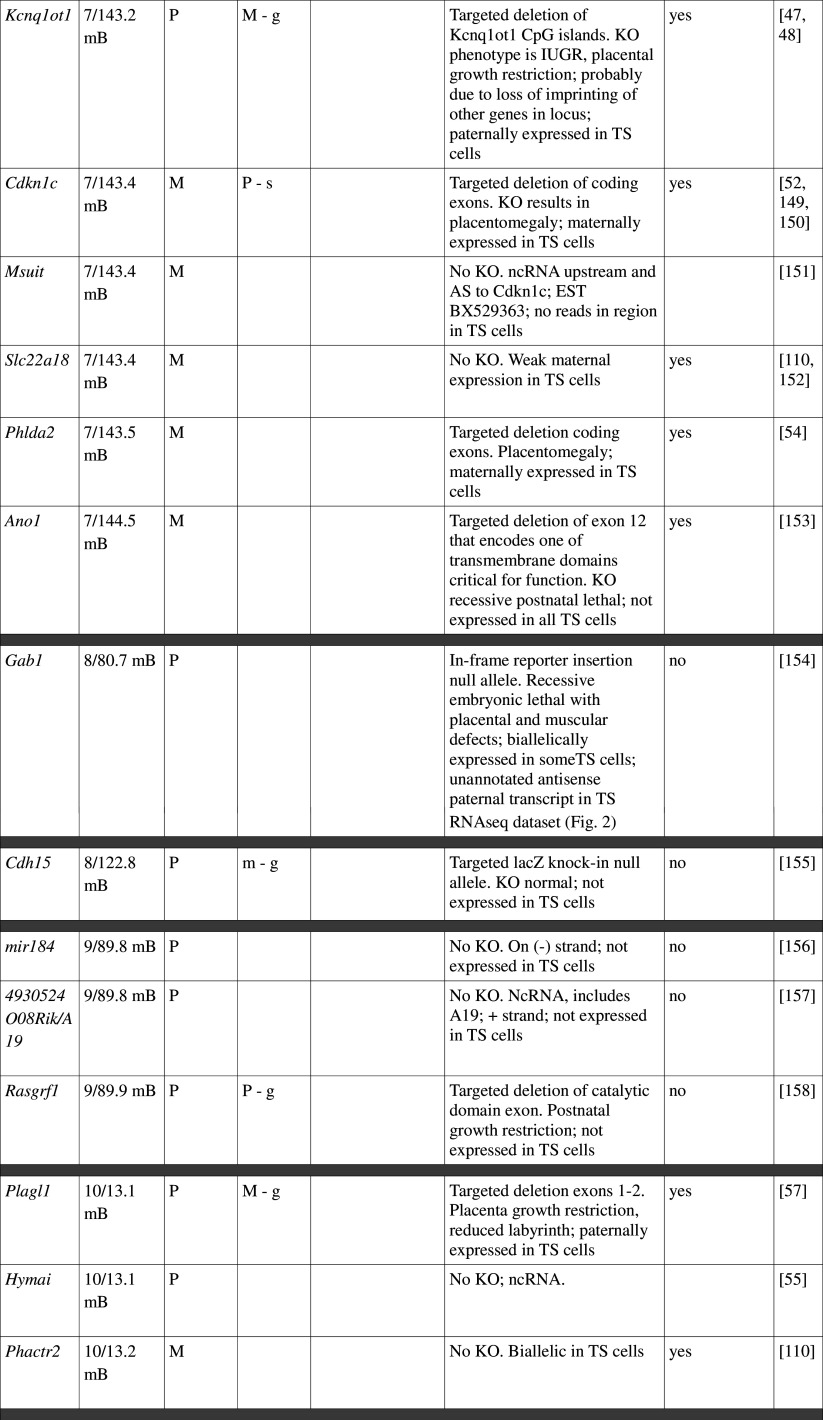
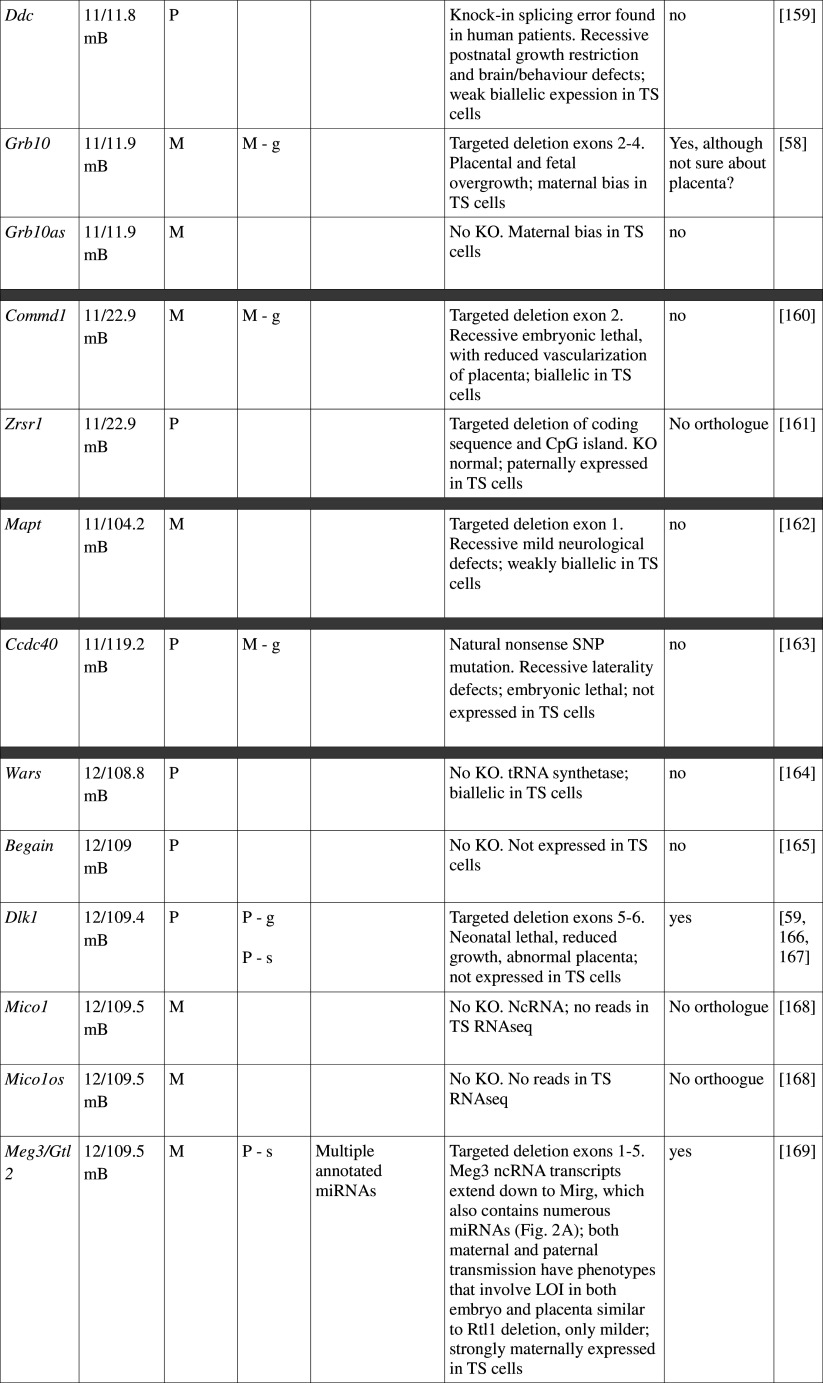
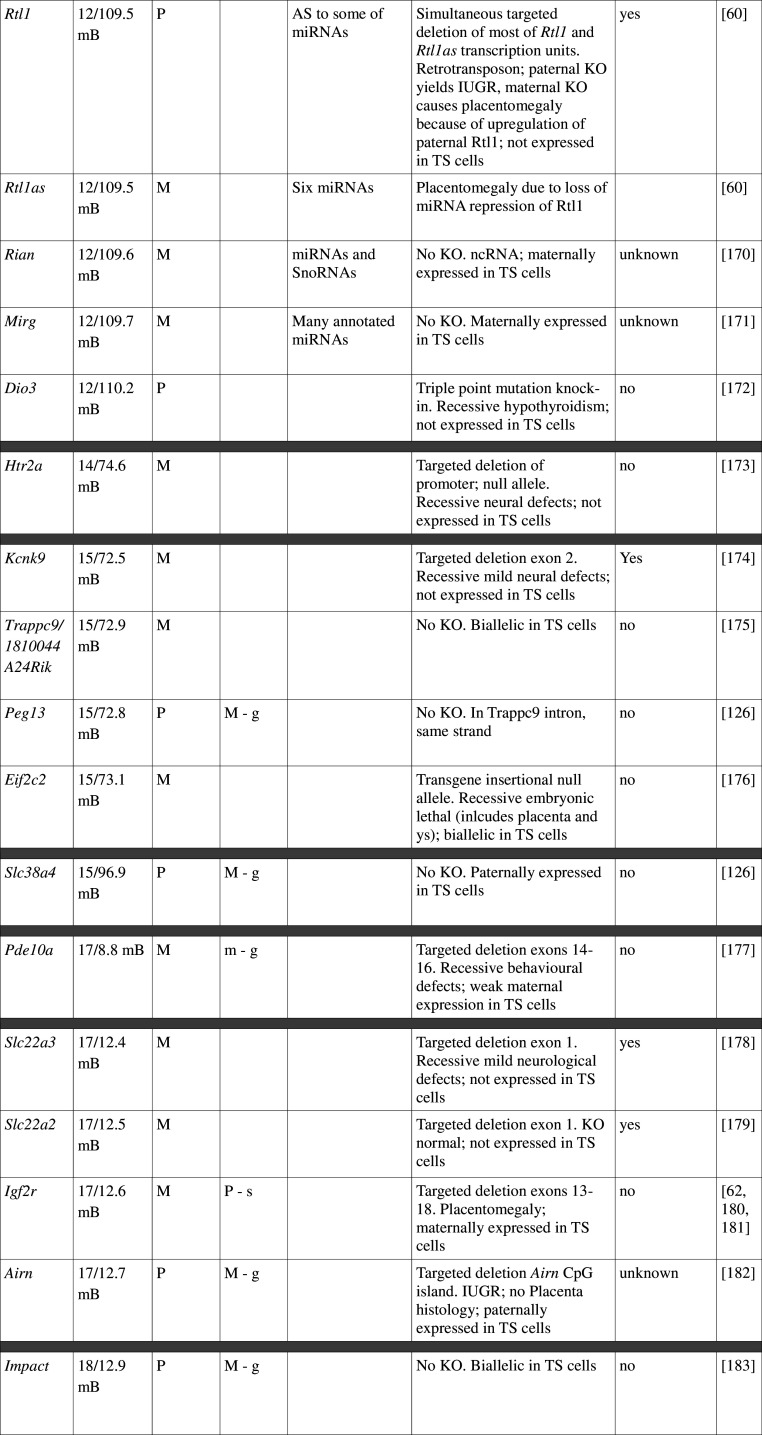
Table 1.
Imprinted gene function
Imprinted genes listed here were obtained from the Otago Catalogue of Parent of Origin Effects website (http://igc.otago.ac.nz/home.html). Where imprinting of a gene is listed as “provisional” or “other”, it has been left out of the table. TS cell expression data were obtained from the dataset published by Calabrese et al. [34]. DMRs with lowercase letters are derived from Kobayashi et al. [74] that have not been confirmed experimentally; “g” denotes germline; “s” denotes somatic. Annotated miRNAs are named unless there are too many. Domains are separated by grey bars