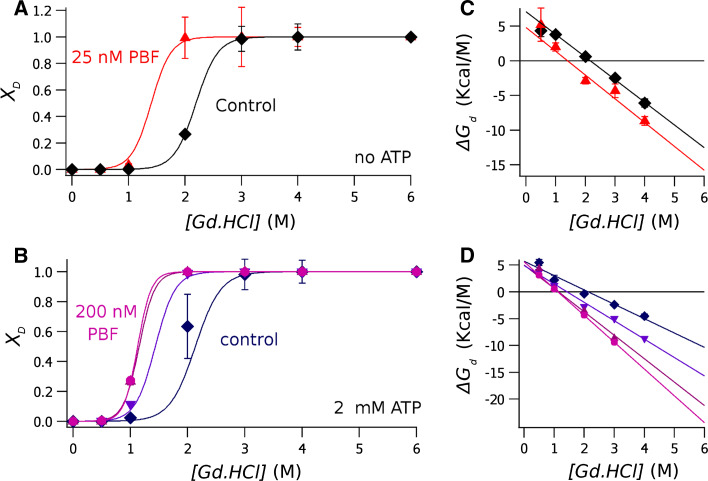
Fig. 3.
Guanidinium denaturation curves of the isomolar mixture of NBD1/NBD2. The molar fraction of the denaturated protein, calculated according to Eq. 3, is plotted against the Gd.HCl concentration. a Data obtained from ATP-free proteins, in absence of PBF (diamonds) and with 25 nM PBF (triangles). b Result deriving from from experiments done in the presence of 2 mM ATP, showing control data (diamonds) and data obtained in the presence of the potentiator at a concentration of 12.5 nM (inverted triangles), 25 nM (triangles) and 200 nM (circles). c, d plots of ΔG d at each Gd.HCl concentration, calculated with Eq. 4 from data shown in (a, b), respectively. Data points are the average of different experiments, and error bars the standard deviation of the mean. The continuous lines were calculated from Eq. 5. The results of the fitting are shown in Table 1