Fig. 16.
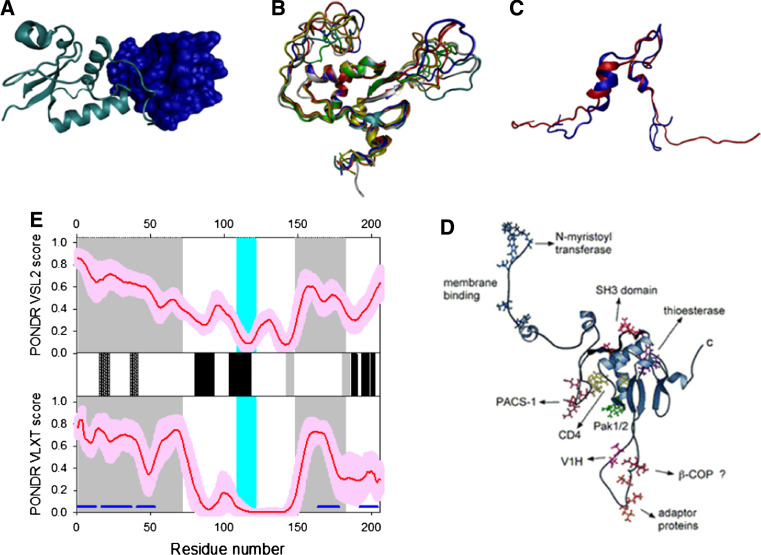
Disorder propensity and structural features of the HIV-1 negative factor (Nef protein). a Crystal structure of the complex between the conserved Nef core (cyan ribbon) and a Src family SH3 domain (blue surface) (1EFN). b NMR solution structure of the unbound Nef core Nef (2NEF). Ten representative members of the conformational ensemble are shown by ribbons of different color. c NMR solution structure of the unbound Nef anchor domain (1QA5). Ten representative members of the conformational ensemble are shown by ribbons of different color. d Structure of HIV-1 Nef protein with some functional annotations discussed in the text. This panel is reproduced with permission from [323]. e Prediction of intrinsic disorder in Nef by PONDR® VSL2 (top panel) and PONDR® VLXT (bottom panel). Red line represents an averaged disorder score for Nef from ~50 different HIV-1 isolates. Pink shadow covers the distribution of disorder scores calculated for Nef from these isolates. Locations of the α-helices seen in NMR structure of the anchor domain are shown by crossed gray bars, whereas gray and black bars between the disorder score panels represent locations of β-strands and α-helices, respectively