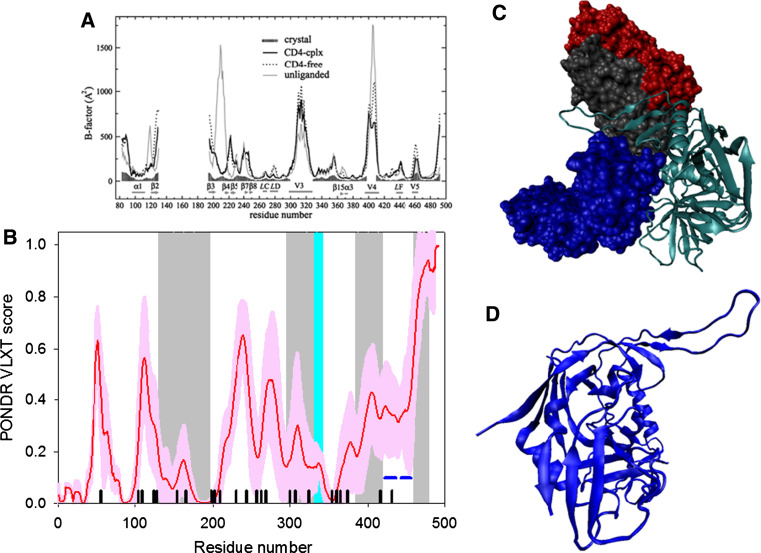
Fig. 4.
Disorder propensity and structural features of the HIV-1 gp120 protein. a Comparison of gp120 Cα B-factors calculated from RMS fluctuations of CONCOORD ensembles between the CD4-complex (solid line), CD4-free (dashed line), and unliganded (gray line) states. The experimentally determined B-factors of the CD4-bound gp120 (PDB entry 1G9M) are shown in gray area. Note that the experimental B-factors of the loops V1/V2, V3, and V4 are missing. The CD4-complexed gp120 is abbreviated to CD4-cplx gp120 and the secondary structure elements and loops V3 and V4 were marked according to the crystal structure of the CD4-bound gp120 [119]. b Disorder prediction evaluated by PONDR® VLXT for the HIV-1 gp120 protein. Red line represents an averaged disorder score for gp120 from ~50 different HIV-1 isolates. Pink shadow covers the distribution of disorder scores calculated for gp120 from these isolates. Gray shaded areas correspond to the mobile loops V1/V2, V3, and VL4. Cyan shaded area shows a CD4 binding loop (residues 332–342). Dark blue lines represent predicted α-MoRFs. c Crystal structure of the HIV-1 gp120 core (light blue ribbon) in complex with a two-domain fragment of CD4 (blue surface) and a neutralizing human antibody dimer (red and gray surfaces) (1GC1). d Crystal structure of the bound form of the HIV-1 gp120 core computationally extracted from its complex with a two-domain fragment of CD4 and a neutralizing human antibody dimer