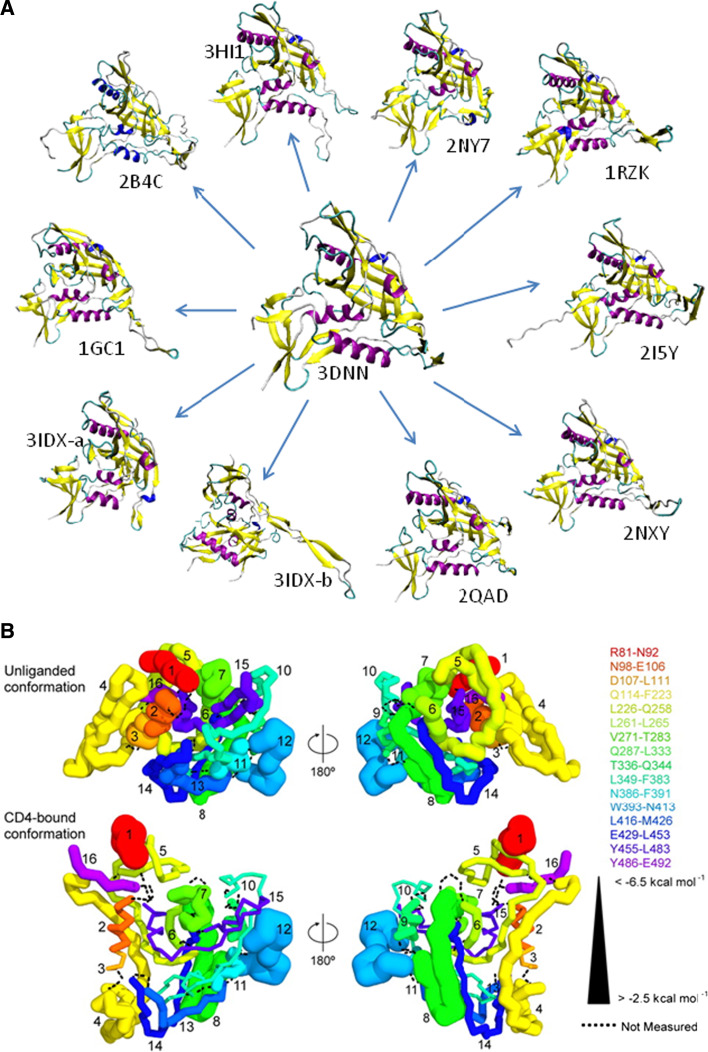
Fig. 5.
Dynamic nature of the HIV-1 gp120 evidenced by the X-ray and hydrogen–deuterium exchange (HDX) experiments. a Multitude of bound conformations of gp120 in complexes with various binding partners. The unliganded gp120 is shown at the center, whereas surrounding structures represent various liganded forms. All structures (except to 3IDX-b) are shown in (relatively) similar orientations to emphasize the structural diversity of the gp120 bound conformations. 3IDX-a and 3IDX-b represent two different orientations of the bound HIV-gp120 core in complex with the Cd4-binding site antibody b13. In 3IDX-b, gp120 is rotated to visualize a long arm used to bind to b13. b Representation of the local gp120 conformational stability derived from the HDX experiments as mapped onto gp120 crystal structure. Combining HDX-determined stability data with atomic-level structural information allows the local conformational stability of gp120 to be visualized. Energies of HIV-1 gp120 conformational stability for 16 peptic fragments are mapped onto a homology model of unliganded gp120 (top row) and onto the CD4-bound crystal structure of YU2 core gp120 (second row). Structures are displayed in Cα-worm representation. Peptic fragments are colored and numbered according to their positions in sequence, with Cα-worm thicknesses corresponding to energies of conformational stability (as shown in the key on the right). Dotted lines indicate regions that were not measured [120]