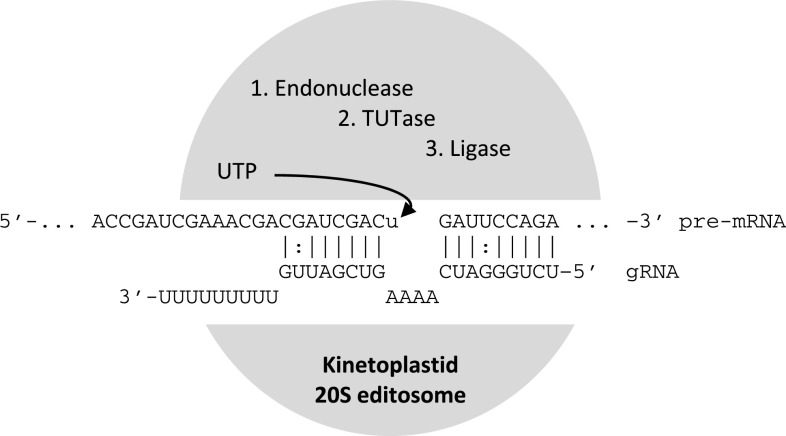
Fig. 2.
A highly simplified view of the uridine insertion type of RNA editing in kinetoplastid mitochondria. Small, 3′-oligo-uridylated antisense guide RNAs (gRNAs) pairing with a given pre-mRNA carry information on location and number of uridines to be inserted. The (entirely hypothetical) example shown displays a case for insertion of four uridines into the pre-mRNA that are ultimately complementary to the initially unpaired adenosines of the gRNA. The different enzymatic activities for RNA editing are assembled in 20S editosome multi-protein complexes, which come in at least three different variants of protein composition, also depending on the location and mode (U insertion vs. deletion) of editing. The three major biochemical activities for uridine insertion are an endonuclease activity cleaving the pre-mRNA at the site of editing, a terminal uridylyl transferase adding uridylates from UTPs to the free 3′OH end of the upstream part of the pre-mRNA, and a ligase rejoining the transcript ends after editing. In the case of uridine deletion editing, a 3′-uridine-exonuclease of the ~20S editosome comes into play that removes unpaired “extra” uridines from the pre-mRNA, which remained unpaired in the hybrid with the respective gRNA