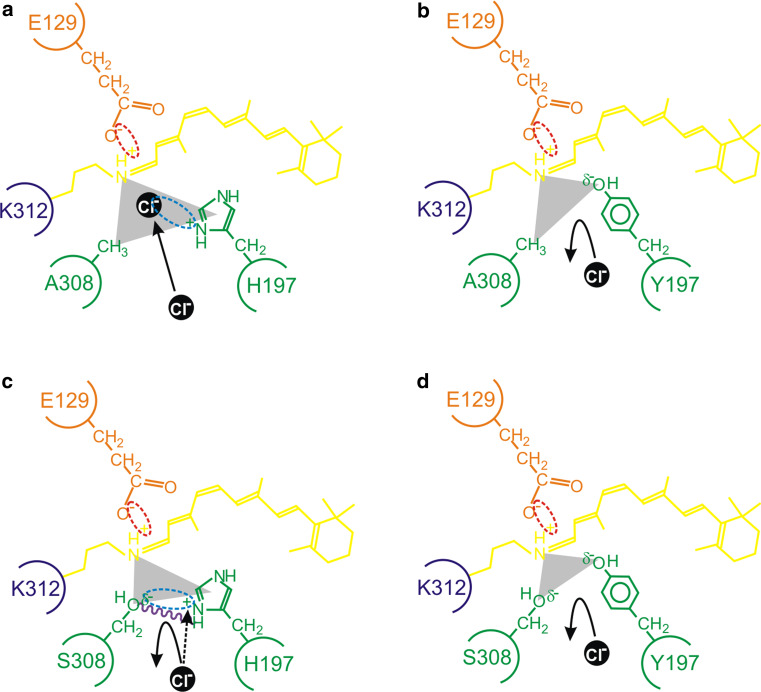
Fig. 3.
A schematic representation of the proposed model for the interaction between residues 197 and 308 (green) in close proximity to the Schiff base (dark blue) of the chromophore (yellow), showing a a chloride ion (black) acting as a counterion (interaction shown by a light blue dotted ellipse) to the positive charge present on H197; b the failure of binding and repulsion of a chloride ion due to a H197Y substitution that results in the loss of a positive charge (from H197) and the gain of a slight negative charge (δ−) (from Y197); c the δ− of the hydroxyl side chain of S308 repelling the negatively charged chloride ion and serving to reduce the strength of the positive charge of H197 (interaction shown by a light blue dotted ellipse); d the complete repulsion and loss of chloride ion binding due to the lack of a positive charge at site 197, the steric hindrance of S308, and the overall increase of a negative charge (δ−) from the side groups of Y197 and S308 (as observed in the wild-type mouse MWS photopigment). In panel c, putative intramolecular hydrogen bond formation (purple wavy line) further restricts the access of a chloride ion. As discussed in the main text, diverse vertebrate species (e.g. the mouse compared to the elephant shark) exhibit subtly different mechanistic roles for chloride ion binding. In some cases (e.g. the elephant shark), the presence of a serine residue at site 308 completely disrupts the chloride-binding site and photopigments generate in the presence or absence of chloride ions. By contrast, the absence of chloride ions in mouse mutant photopigments possessing H197 (whether site 197 is occupied by an alanine or serine residue) fail to generate successfully. This suggests that a chloride ion that should be repelled from binding directly within the binding pocket may exert some influence over protein structural fidelity and may partly act as a counterion (black dotted arrow) to the positive charge of H197 from a position that is external to the regular location of the chloride-binding site. In all cases, the E129 counterion (orange), the Schiff base linkage at K312 (dark blue), the retinylidene chromophore (yellow) and the triform spatial representation of the chloride-binding pocket (grey scalene triangle) are shown. Electrostatic attractions are highlighted between the side chain of E129 and the Schiff base (red dotted ellipse), the side chain of a histidine residue present at site 197 and a chloride ion (light blue dotted ellipse), and the amino acid side chains of H197 and S308 (light blue dotted ellipse)