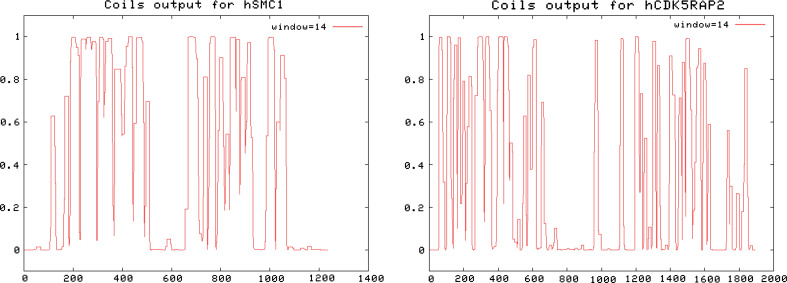
Fig. 3.
Predicted coils in human SMC1 and CDK5RAP2. Coiled coil regions were predicted in (a) human SMC1 and (b) human CDK5RAP2 proteins using the corresponding EMBnet program (http://www.ch.embnet.org/software/COILS_form.html) according to Lupas et al. [11]. COILS is a program that compares a sequence to a database of known parallel two-stranded coiled-coils and derives a similarity score. By comparing this score to the distribution of scores in globular and coiled-coil proteins, the program then calculates the probability that the sequence (amino acid position represented in x-axis) will adopt a coiled-coil conformation (probability 0–1 represented in y-axis). A comparison of the predicted conformation of human SMC1 (structural maintenance of chromosomes protein 1A, NP_006297.2) and human CDK5RAP2 (NP_060719.4) reveals structural similarities between the two proteins. Both are composed of two long coiled-coil motifs connected by a rather unstructured domain that in the case of SMC proteins has been predicted to act as a flexible hinge that allows the proteins to gain a V-shaped structure necessary for the C-and N-terminal domains to come in close contact. The settings of the coils program were set to default, using a window width of 14, 21 and 28 amino acids