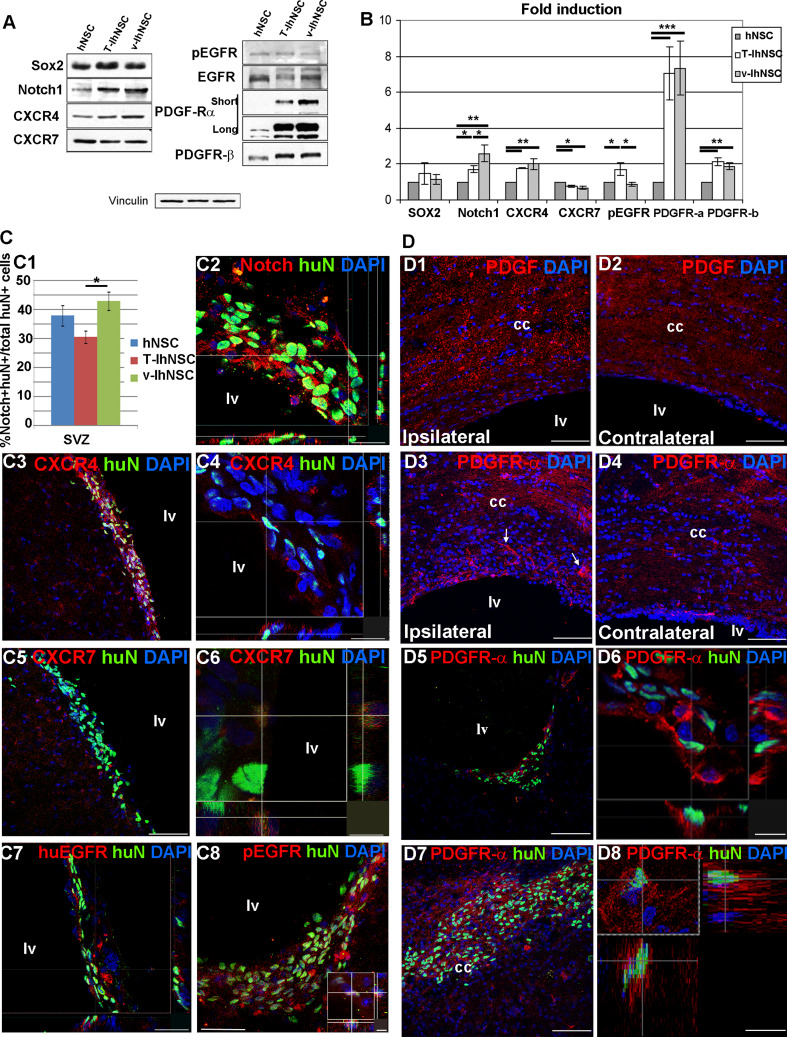
Fig. 3.
Candidate mechanisms for hNSC, T-IhNSC, and v-IhNSC differential migration: Western-blot (a) and relative quantification (b) of Sox2, notch1, CXCR4, CXCR7, pEGFR, EGFR, PDGFαR, and PDGFβR in hNSC-, T-IhNSC-, and v-IhNSC-derived progenitors. c Immunohistochemistry showing the distribution and colocalization with huN+ of putative proteins involved in the integration of transplanted cells into the SVZ. c1 Chart showing the percentages of huN+/notch1+ cells over total huN+ cells and c2 confocal image of huN+/notch1+ cells in the ventricular wall. Co-localization of CXCR4 (c3, c4), CXCR7 (c5, c6), huEGFR (c7), and pEGFR (c8 and inset) with huN+ T-IhNSC in the SVZ. d Putative proteins involved in the migration of transplanted cells to the lesioned area. Distribution of endogenous PDGF ligand (d1–d2) and PDGFαR (d3–d4), respectively, in the lesioned (d1, d3) and contralateral (d2–d4) hemisphere. Note PDGFR-α+ cells migrating to the lesioned cc (arrows d3). Co-localization of PDGFαR with huN in lv (d5, d6) and in cc (d7, d8). Total nuclei are shown by DAPI staining (blue). Scale bars, in c1, c4 15 μm, in c2, c7 23 μm, in c3, c5, d1–d5, d7 75 μm, c6 7 μm, c8 37 μm (inset 5 μm) and d6, inset, d8 12 μm. lv lateral ventricle and cc corpus callosum