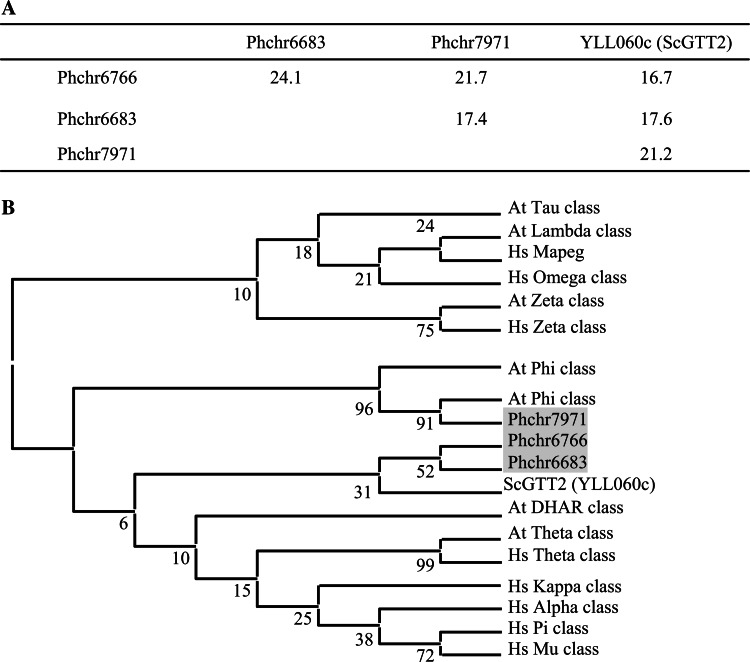
Fig. 1.
GTT2-related sequence comparisons. a Percentage of identity between GTT2-related sequences from P. chrysosporium and S. cerevisiae (ScGTT2), based on global alignment determined by Lalign software. (http://www.ch.embnet.org/software/LALIGN_form.html). b Phylogenetic distribution of GTT2-related sequences of P. chrysosporium among GST classes from Homo sapiens, A. thaliana, and S. cerevisiae. The diagram has been drawn based on a phylogenetic analysis carried out with ClustalW and MEGA4 software. The analyses were conducted using the neighbor-joining (NJ) method implemented in MEGA, with the pairwise deletion option for handling alignment gaps, and with the Poisson correction model for distance computation. Bootstrap tests were conducted using 1,000 replicates. Branch lengths are proportional to phylogenetic distances. Sequences from human, A. thaliana and S. cerevisiae were obtained from NCBI databases (http://www.ncbi.nlm.nih.gov/). Sequences from P. chrysosporium were obtained from JGI (http://genome.jgi-psf.org). Hs Homo sapiens, At A. thaliana, Sc S. cerevisiae and Pc P. chrysosporium. Accession numbers for A. thaliana proteins are: At5g41210; At5g41240; NP_198938; X68304; At4g02520; At2g02930; At1g02950; At1g02940; At1g02930; At1g02920; At2g47730; At2g30860; At2g30870; At3g03190; At5g17220; At3g62760; At1g49860; Q9ZVQ3; Q9ZVQ4; At2g29490; At2g29480; At2g29470; At2g29460; At2g29450; At2g29440; At2g29420; At3g09270; At5g62480; At1g74590; At1g69930; At1g69920; At1g27130; At1g27140; At1g59670; At1g59700; At1g10370; At1g10360; At1g78380; At1g78370; At1g78360; At1g78340; At1g78320; At1g17170; At1g17180; At1g17190; At3g43800; At1g53680; At5g02790; At5g02780; At3g55040; and for human proteins are: AAB96392; AAC13317; AAA60963; AAA70226; NP_899062; NP_665735; CAA33508; Q9Y2Q3