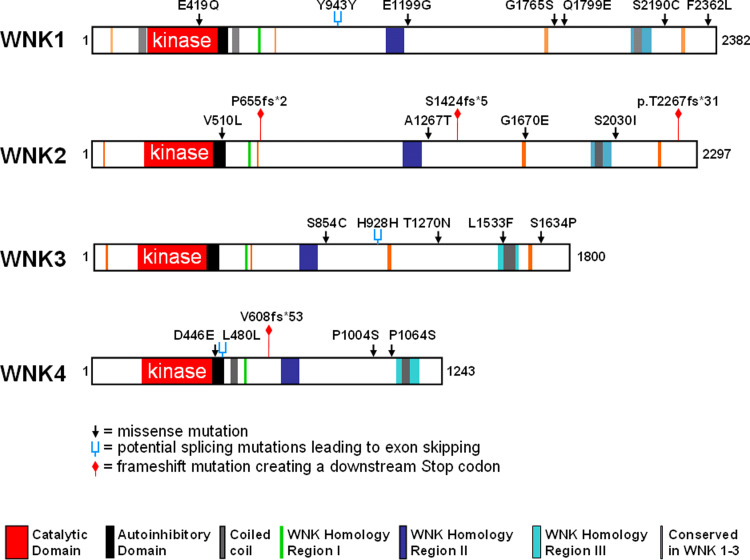
Fig. 4.
Graphic representation of the somatic WNK mutations described in Table 2. The positions of the mutations are illustrated with respect to the catalytic, autoinhibitory and coiled-coil domains. Missense mutations are given as black arrow heads and frameshift mutations leading to premature stop codons are marked by red signposts. Silent nucleotide changes are marked as blue forks because they can act at the nucleotide level by interfering with exon recognition during the pre-mRNA splicing process and result in exon skipping. A sequence analysis of the three silent changes using the ASD database (http://www.ebi.ac.uk/asd-srv/wb.cgi?method=8) indeed indicates a potential loss of splicing factor binding sites, namely for SC35 in WNK1 Y943Y, for ASF/SF2 and SC35 in WNK3 H928H, and for SRp40 in WNK4 L480L