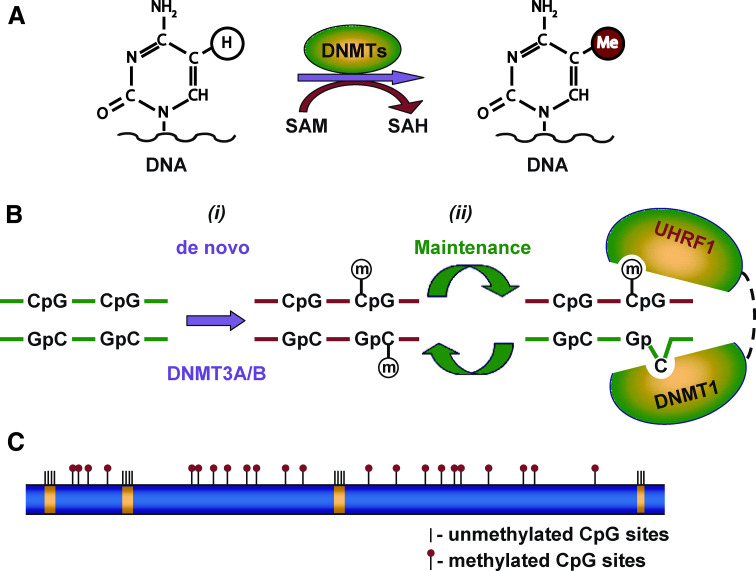
Fig. 1.
Schematic model showing cytosine DNA methylation. a Cytosine residues in DNA at CpG sites are converted 5-methylcytosines by the addition of a methyl group from SAM to the fifth carbon atom in the cytosine pyridine ring. This reaction is catalyzed by the enzymatic activity of DNA methyltransferases (DNMTs). b Establishment and maintenance of the DNA methylation pattern during DNA replication. (i) DNA methylation is initiated and established during embryonic development by means of the de novo DNMT3A and DNMT3B DNA methyltransferases. (ii) Maintenance of DNA methylation. During DNA replication, DNA methylation is maintained by a complex coordinated action of the maintenance methyltransferase DNMT1 and UHRF1 [28–30]. The SRA domain of UHRF1 recognizes the hemimethylated CpG site and recruits DNMNT1, which transfers methyl group to the unmethylated cytosine residue on the newly synthesized DNA strand. c DNA methylation landscape in mammalian genomes. Methylation in normal mammalian cells occurs primarily at CpG sites located in repetitive sequences, exons other than first exons, and intergenic DNA (blue). The CpG islands that span the promoter and first exons of the majority of genes are usually unmethylated (yellow) in normal cells and embedded in a matrix of long methylated domains (blue) [33, 34]