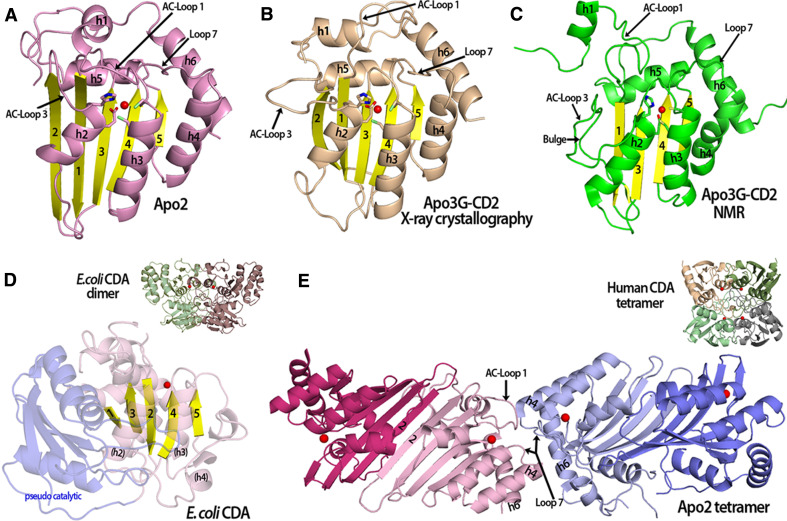
Fig. 1.
Comparison of APOBEC and fntCDA structures. a An Apo2 monomer (PDB 2nyt, chain A-closed loop conformation) showing the catalytic residues and a zinc molecule (red sphere) in the active center. b The crystal structure of an Apo3G-CD2 monomer (PDB 3e1u) contains 5 beta strands similar to Apo2 and other deaminases. c An Apo3G-CD2 structure solved by NMR (PDB 2KBO) which is very similar to the other APOBEC structures (see a and b). In contrast to the Apo3G-CD2 crystal structure, the NMR structure has a discontinuous beta 2 strand that contains a bulge. Also, AC-Loop 1 and h1 are positioned differently in comparison to the Apo3G-CD2 structure in (b). d The structure of an ECDA monomer containing a pseudo-catalytic domain (light blue) connected to the catalytic domain (light purple with yellow beta strands) via a long flexible loop. The active center zinc is represented as a red sphere. Unlike the APOBEC structures that contain a long h4 and h6, the equivalent ECDA h4 forms a long flexible loop and small 310 helices that connect to the pseudo-catalytic domain (inset). The square-shaped E. coli CDA dimer (PDB 1ALN) with the monomers shown in green and purple, each containing an active center zinc (red sphere). e The Apo2 tetramer (PDB 2nyt) displays a novel type of oligomerization forming dimers through a head–head interaction. H4 and h6 allow for the extended tetramer formation with interactions occurring though residues from Loop 7, AC-Loop 1, h4 and h6. The closed AC-Loop 1 conformations found in the inner monomers (light pink and light blue) at the tetramer interface may block substrate access to these active centers. Each monomer is colored differently with a zinc ion in the active center (red sphere) (inset). The human CDA square-shaped tetramer (PDB 1mq0) with the monomers shown in different colors and the zinc ion represented as a red sphere