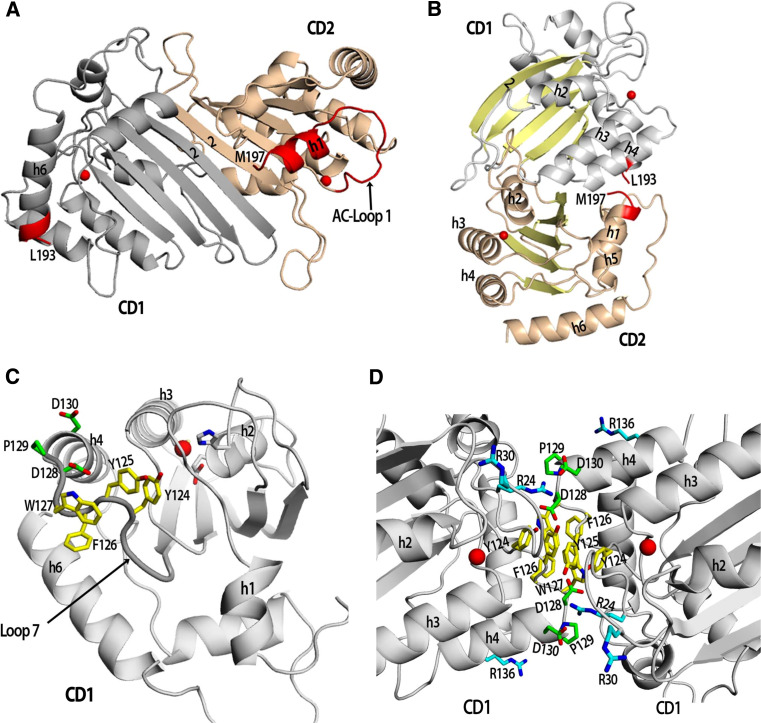
Fig. 3.
Models of Apo3G monomers and dimers. a A model of an Apo3G monomer whereby the two domains fold similar to an Apo2 dimer by pairing of the β2 strands. The inter domain connection between Apo3G-CD1 and Apo3G-CD2 would have to be made through an additional three amino acids (194–196) that could connect Apo3G-CD1 h6 (L193) to h1 of Apo3G-CD2 (M197) (connection to be made is highlighted in red). The SWISS-MODEL protein homology modeling program was used to generate the Apo3G-CD1 model using an Apo2 monomer in a closed conformation as a template [106]. PyMOL was used to generate the full-length Apo3G monomer using an Apo2 dimer as a template (PDB 2nyt): the Apo3G-CD1 model was aligned with one Apo2 monomer and the structure of Apo3G-CD2 (PDB 3e1u) with the other Apo2 monomer [107]. Apo3G-CD1 is colored grey, Apo3G-CD2 is colored wheat and the two zinc molecules are represented by red spheres. b A different model of an Apo3G monomer adapted from Harjes et al. where the two active center domains of Apo3G-CD1 and Apo3G-CD2 lie on opposite faces (zinc ion represented by red spheres). The Apo3G-CD2 structure (3e1u) was aligned with the Apo3G-CD1 model according to Harjes et al. [5]. c A model of Apo3G-CD1 displaying the residues on Loop 7 important for dimerization (yellow residues) and Vif binding (green residues). d A model of the proposed Apo3G dimer CD1–CD1 (head–head) interface based off the Apo2 tetramer interface (PDB 2nyt). Yellow colored residues (Y124, Y125 and W127) have been proposed to be important for dimerization and virion incorporation. The residues (D128, P129 and D130) that interact with Vif are colored green. Residues important for binding RNA, R24, R30 and R136, are colored cyan