Fig. 2.
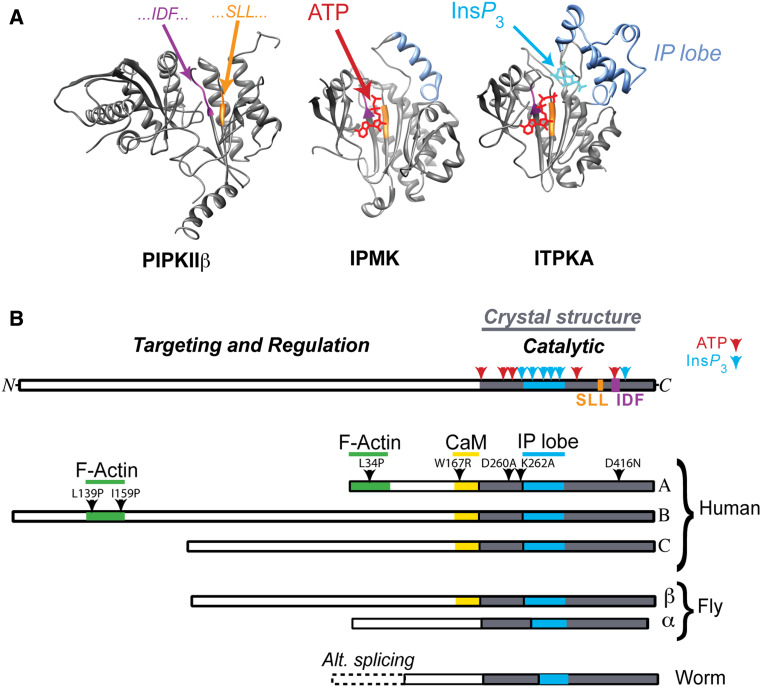
Structure of ITPKs. a Comparison of crystal structures of kinase superfamily members. Crystal structures of human phosphatidylinositol 4-phosphate 5-kinase beta (PIPKIIβ, left) [212], yeast inositol polyphosphate multikinase (IPMK, middle) [27], and human ITPKA (right) [21]. Structures are oriented such that one is looking into the active sites, which bind ATP (red) through a conserved motif (SLL…IDF…, colored purple/orange). Inositol phosphates (IPs), such Ins(1,4,5)P 3 (InsP 3, blue) are oriented through their interaction with the IP lobe (blue), which holds the IP so that phosphotransfer from ATP is facilitated. IPMK (middle) has a smaller IP lobe than ITPK, allowing it to accommodate multiple IP substrates; by contrast, the more elaborated IP lobe found in ITPKs (right) holds Ins(1,4,5)P 3 in an orientation that allows phosphorylation at the 3-position only. b Domain structure of the three human ITPK isoforms compared to fly and worm varieties. The catalytic domains of ITPKs are highly conserved and located in the C-terminal parts of the protein (top domain diagram). By contrast, the amino terminal domains are divergent, and govern selective targeting and regulation in different cells and tissues. All ITPK crystal structures solved thus far have been obtained from catalytic fragments (gray), which lack the N-terminal regulatory regions; the catalytic region contains all of the substrate binding sites for ATP (red) and InsP 3 (blue). In the presence of Ca2+, all three human isoforms bind calmodulin (CaM) in a Ca2+-dependent manner, via a conserved domain [45]. The three human isoforms of ITPK show diversity in their N-termini [8]. Isoforms A [177] and B [77] bind filamentous actin (F-actin), while isoform C shows a mixed cytosolic/nuclear localization [213]. Arrowheads are labeled with point mutations that can selectively destroy domain functions in ITPKA or ITPKB. The L34P mutation destroys F-actin binding in ITPKA [133], while L139P and L159P reduce F-actin binding in ITPKB [77]. The W167R mutation destroys CaM binding in ITPKA; analogous mutations behave similarly for ITPKB and C [45]. Arrowheads in the blue and gray areas point to two of the many mutations in the active site that destroy catalytic activity [21]. The fruit fly Drosophila melanogaster possesses two isoforms [42], one of which is positively regulated by calmodulin [45]. The nematode Caenorhabditis elegans posesses one ITPK gene, which is not regulated by CaM and shows diversity generated by alternative splicing at the 5′ end [39]