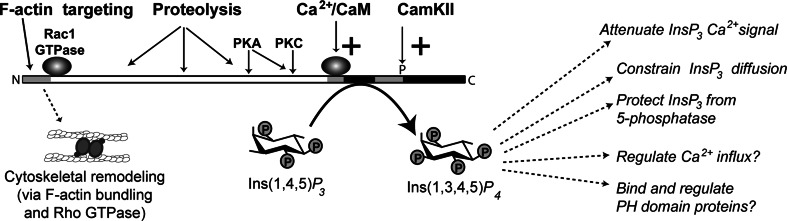
Fig. 3.
Regulation of ITPKs and its consequences. ITPK action is regulated by at least four mechanisms. All mammalian ITPKs are positively regulated by Ca2+/calmodulin; this creates a negative feedback loop on the Ins(1,4,5)P 3 signal to release Ca2+ via ITPR [45]. ITPKs are also regulated by various protein kinases, such as protein kinase C (PKC), protein kinase A (PKA), and Ca2+/CaM-activated protein kinase II (CamKII). Phosphorylation at some sites activates the enzyme, and at others inhibits it (see text). The amino terminal regions of ITPKA and ITPKB bind F-actin, which targets the enzymes near sites of Ins(1,4,5)P 3 generation. The ITPKA amino terminal-binding region [177] affects F-actin structure through a mechanism independent of ITPKA catalytic activity [179]. This mechanism has been linked to the Rho family GTPase Rac1 [132]. The F-actin remodeling involves cross-linking or bundling of the actin filaments, and controls the selective targeting of ITPKA to dendritic spines [133]. ITPKs are subject to the actions of proteases, which usually cleave between the N-terminal targeting region and the catalytic region. Protease cleavage of ITPKs changes enzymatic localization [74, 75], and can also affect the sensitivity to or substrate sites for protein kinases [70]. ITPK catalysis has many suggested functions in cells [10]. These can be subdivided into functions involving the attenuation of Ins(1,4,5)P 3 signals via restriction of those signals to spatial and temporal domains through the control of Ins(1,4,5)P 3 lifetime, and those which depend on the specific actions of the enzymatic product Ins(1,3,4,5)P4. Cells possess a variety of Ins(1,3,4,5)P 4-binding proteins. Some of these bind Ins(1,3,4,5)P 4 selectively via their pleckstrin homology (PH) domains; others are channel proteins whose gating or conductance properties change upon Ins(1,3,4,5)P 4 binding (see text)