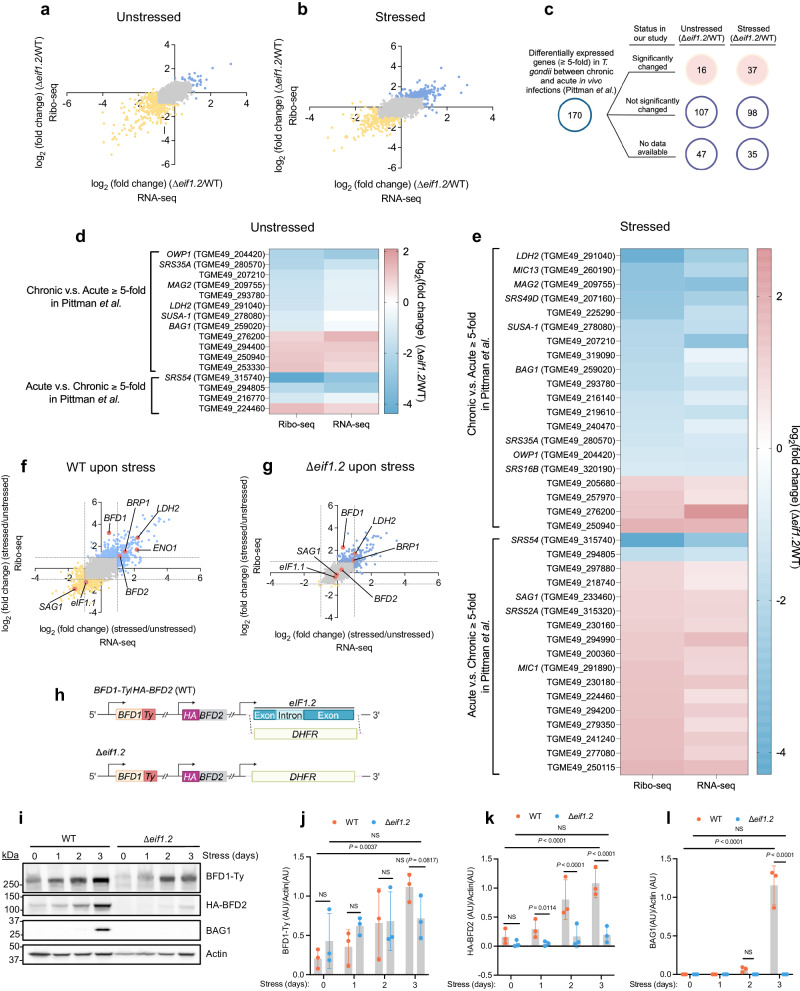
Fig. 5. eIF1.2 regulates the expression of stage-specific factors, including BFD1 and BFD2.
a–g The impact of eIF1.2 deletion on transcription and translation. RNA-seq, RNA sequencing. Ribo-seq, Ribosome profiling. n = 3 biological replicates. a, b, f, g Blue dots, genes increased for at least 2-fold (Padj < 0.05). Yellow dots, genes decreased for at least 2-fold (Padj < 0.05). Padj values were derived using DeSeq2, employing a two-sided test with adjustments for multiple comparison. Grey dots, not significantly changed genes. Pink dots, genes of interest. Grey dotted lines indicate where the y-axis or x-axis equals 1 or −1. Stressed, 1 day of alkaline stress. a Comparison of unstressed ∆eif1.2 and WT (ME49∆ku80) parasites at transcription and translational levels. b Comparison of stressed ∆eif1.2 and WT (ME49∆ku80) parasites at transcription and translational levels. c–e Comparing our RNA-seq and Ribo-seq results with a RNA-seq dataset for in vivo acute and chronic infection17. Listed significantly changed genes in our study have a fold change greater than 2 or less than 0.5 in either RNA-seq, Ribo-seq or both, with Padj < 0.05, and minimum of 5 reads. d Heatmap illustrates the fold change of the 16 genes that were significantly altered between unstressed ∆eif1.2 and WT parasites in our study (Padj < 0.05). e Heatmap illustrates the fold change of the 37 genes that were significantly altered between stressed ∆eif1.2 and WT parasites in our study (Padj < 0.05). f The impact of alkaline stress on transcription and translation in WT parasites. g The impact of alkaline stress on the transcription and translation in ∆eif1.2 parasites. h Knocking out eif1.2 in a strain24 with both BFD1 and BFD2 tagged. i Western blot analysis of lysates from parasites exposed to alkaline stress for indicated days. Actin was used as a loading control. j–l Quantification of western blot results. Data represent the mean ± s.d. n = 3 biological replicates. The ratios of BFD1-Ty/Actin, HA-BFD2/Actin and BAG1/Actin were analyzed using a linear mixed model. NS, not significant (P > 0.05). Source data are provided as a Source Data file.