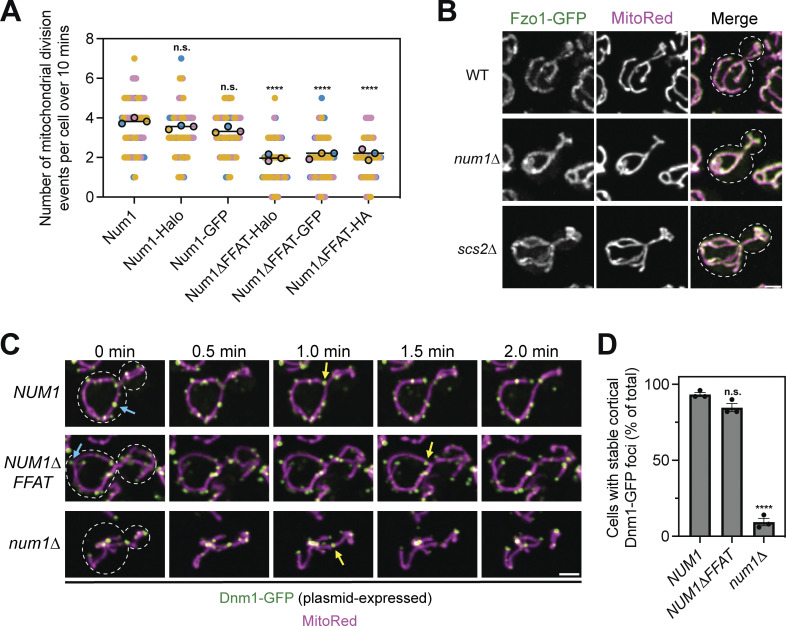
Figure S3.
Loss of Num1 does not alter the localization of the mitochondrial division or fusion machinery. (A) The presence of a fluorescent or epitope tag on Num1 does not influence the rate of mitochondrial division. Imaging and quantification of mitochondrial division was performed identically to (Fig. 4 B), except cells were expressing the indicated Num1 or Num1∆FFAT fusion proteins. The data from Num1-Halo and Num1∆FFAT-Halo are reproduced from (Fig. 4 B) to aid visual comparison. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistical analyses are in comparison to Num1-Halo. (B) Fluorescent micrographs of cells expressing MitoRed and genomically tagged Fzo1-GFP in wild type, num1∆, and scs2∆ backgrounds. Individual channels are shown in grayscale. Dashed white lines indicate cell outlines. Scale bar, 2 µm. (C) Time-lapse images of cells expressing MitoRed and Dnm1-GFP, expressed from a plasmid, in the indicated genetic backgrounds. The blue arrows point to stable Dnm1-GFP foci that remain cortically localized throughout the time-lapse. The yellow arrows indicate accumulations of Dnm1-GFP on the mitochondria immediately prior to a division event. Images are max projections of full Z-stacks. Scale bar, 2 µm. (D) Quantification of the percent of cells containing at least one cortically localized Dnm1-GFP focus. Cells expressed Dnm1-GFP as an extra copy from a plasmid under control of the native promoter. To be counted, Dnm1-GFP foci needed to remain cortically localized for at least 2 min. Each dot represents one imaging replicate containing 50 cells. Error bars represent SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistical analyses are in comparison to NUM1.