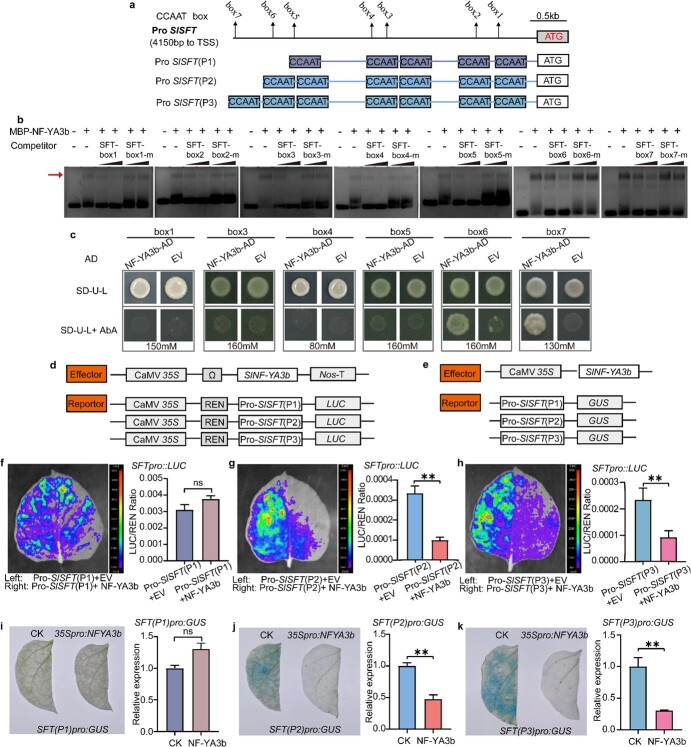
Figure 5.
Specific binding of SlNF-YA3b to the CCAAT cis-elements of the SFT promoter. a Schematic diagram of the 4150-bp SFT promoter region. Seven CCAAT (boxes 1–7) cis-elements were identified in the 4150-bp fragment of the SFT promoter. The translation start codon (ATG) is indicated. TSS, translation start site. Pro SlSFT (P1), Pro SlSFT (P2), and Pro SlSFT (P3) constitute a genomic fragment of the SFT promoter containing boxes 1–5, boxes 1–6, and boxes 1–7, respectively. They are 3589, 3627, and 4150 bp in length upstream of the TSS. b EMSAs for binding of NF-YA3b to the CCAAT cis-elements in boxes 1/2/3/4/5/6/7 of the SFT promoter. NF-YA3b protein was incubated with FAM-labeled box 1/2/3/4/5/6/7 probes, and the mobilities of the protein–oligonucleotide probe complexes on non-denaturing gels are indicated with red arrows. The specific unlabeled competitors (SFT-box1/2/3/4/5/6/7) or unlabeled mutant oligonucleotide competitors (SFT-box1/2/3/4/5/6/7-m) were added to the incubation mixtures of lanes 3, 4, 5, and 6, by a 10- and 30-fold molar excess. −, absence; +, presence. c Y1H experiments for the binding of NF-YA3b to the CCAAT cis-element at different positions of the SFT promoter. Seven constructs containing individual CCAAT cis-elements were used in the assays. The negative control was an empty pGADT7 vector (EV). d Dual-luciferase reporter assays. NF-YA3b was expressed from pGreenII 62-SK under the CaMV 35S promoter and served as an effector. Pro SlSFT (P1), Pro SlSFT (P2), and Pro SlSFT (P3) were cloned into the pGreenII 0800-LUC vector and used to drive the expression of the LUC reporter. e GUS gene expression assays. The full-length CDS of NF-YA3b was cloned into pHellsgate8 for expression of NF-YA3b protein as the effector. Three SFT promoter fragments of 3589 bp (SFT-P1), 3627 bp (SFT-P2), and 4150 bp (SFT-P3) upstream of the translation start codon (ATG) were cloned into pMV2-GUS vector to drive GUS gene expression. f–h Representative images of luciferase activity (left) and ratios of LUC/REN activities (right) in N. benthamiana leaves. Note that SlNF-YA3b suppresses its transcriptional activity on SlSFT (P2) and SlSFT (P3) but does not change its transcriptional activity on SlSFT (P1). The values displayed are mean ± standard error (n = 8). ns, not statistically significant; **P < 0.01 (t-test). i–k Representative images of GUS activity staining (left) and relative GUS gene expression levels (right) in N. benthamiana leaves. In the control (CK), the empty pHellsgate8 vector was used to replace the effector for co-expression with SFT(P1)pro:GUS (i), SFT(P2)pro:GUS (j), and SFT(P3)pro:GUS (k) in N. benthamiana leaves. The GUS gene expression level in the control (CK) was set as 1.0. Data are mean ± standard error (n = 3). ns, not statistically significant, **P < 0.01 (t-test).