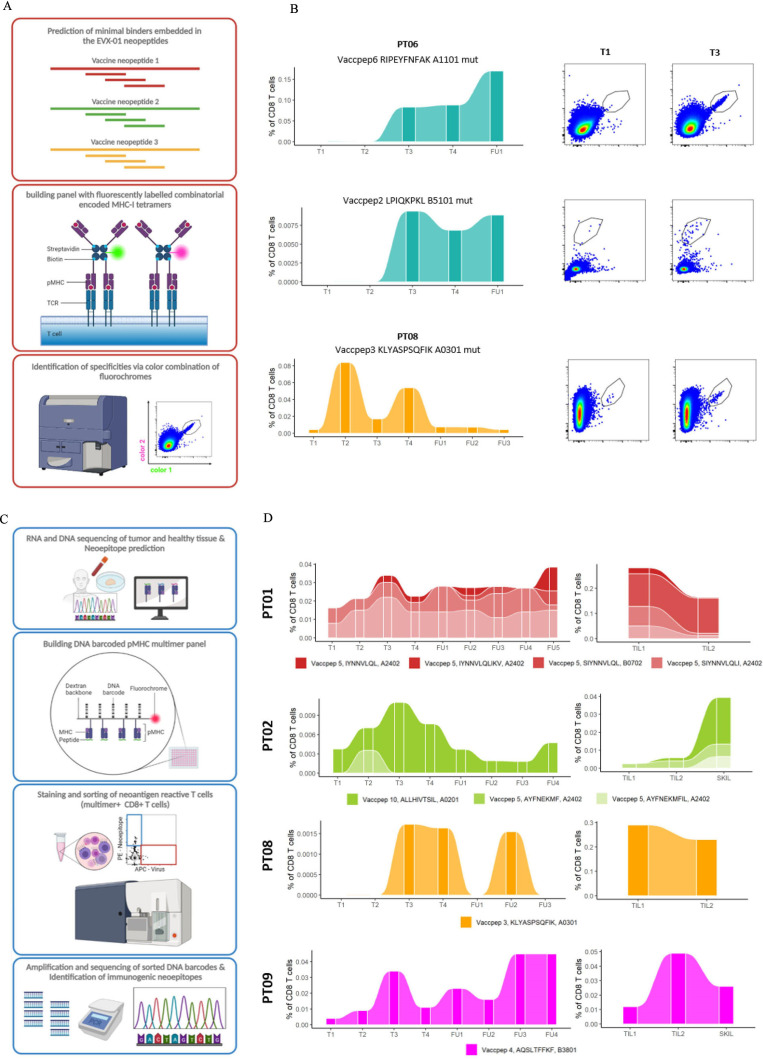
Figure 4.
EVX-01 prestimulated PBMCs and ex vivo PBMC derived CD8+T cells screened for EVX-01 and other CD8+T cell neoepitopes. (A) A schematic overview of the experimental setup for detection of VaccNARTs in prestimulated PBMCs. Minimal peptides embedded in the EVX-01 neopeptides were predicted and loaded on tetramers conjugated to fluorochromes. Specific combinations of two fluorochromes were used for identification of the specificities of detected VaccNARTs. (B) The frequency of selected VaccNART with only one specificity, from patients 6, 8 and 10. EVX-01 peptide number (Vaccpep), short peptide sequence, HLA, and whether the short peptide includes the mutated region of the Vaccpep (mut) is shown above the graphs. dotplots for time point T1 (baseline) and T3 (after three IP vaccinations) are shown for each peptide. (C) A schematic overview of experimental setup used for detection of NARTs, VaccNARTs and VARTs in ex vivo PBMCs, expanded TILs and SKILs. CD8+T cells reactive toward predicted neoepitopes, including vaccine embedded neoepitopes, and virus epitopes were screened with DNA barcoded pMHC multimers. (D) The frequency VaccNARTs shown for selected patients, showing the single populations dynamics over time. EVX-01 peptide number (Vaccpep), short peptide sequence and HLA is shown next to the graphs. IP, intraperitoneal; PBMC, peripheral blood mononuclear cell.