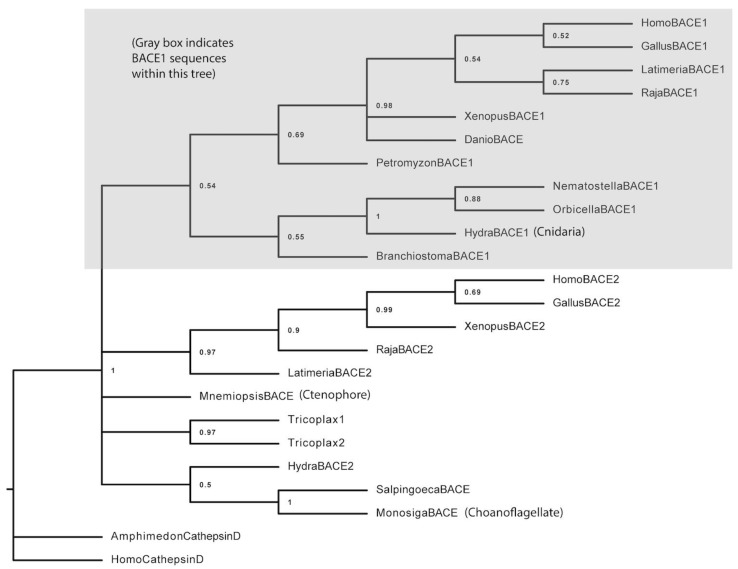
Figure 1.
Cnidarians are the most basal group with a BACE1 ortholog. Maximum-likelihood tree of BACE sequences with nodes collapsed below the 50% bootstrap support. BACE1 and BACE2 sequences form clear clades with cnidarians being the most basal group found in our search to have a BACE1 sequence (shown in the gray box). Other BACE-like sequences can be found in basal animal taxa such as ctenophores (e.g., Mnemiopsis) and placozoans (Tricoplax), as well as in the sister group to animals, choanoflagellates (Monosiga). These sequences form a polytomy with the BACE1 and BACE2 groups, mirroring the polytomies that are found in phylogenies at the whole-organism level. As a tentative model, we consider them to be single-gene, pre-duplicate precursors to BACE 1 and BACE 2. Tricoplax appears to have undergone an independent duplication of this BACE1/2 precursor. Cathepsin D aspartyl proteases are the sister group to the BACE genes; we can find this gene but not BACE genes in porifera (Amphimedon).