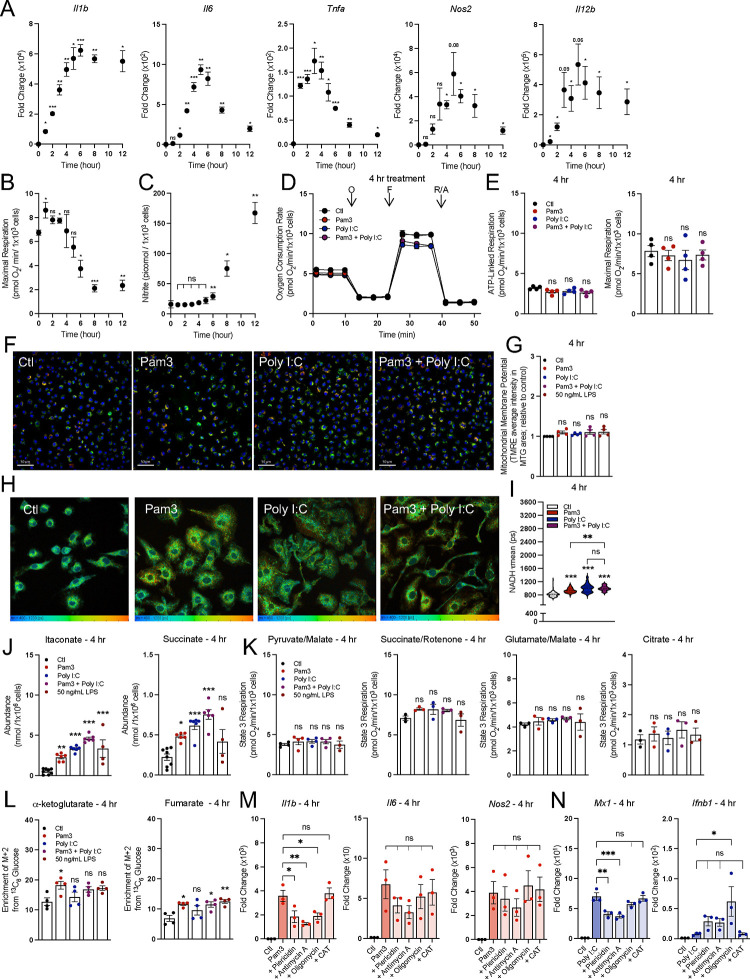
Figure 6: The induction of the pro-inflammatory response does not temporally align with respiratory inhibition.
A) Pro-inflammatory gene expression in control (Ctl) and BMDMs treated with 50 ng/mL LPS across multiple timepoints relative to control (Ctl) (n = 3–4). B) Maximal respiration rate for control (Ctl) and BMDMs treated with 50 ng/mL LPS across multiple timepoints (n = 3–4). C) Levels of nitric oxide in medium from control (Ctl) and BMDMs treated with 50 ng/mL LPS across multiple timepoints (n = 3–4; not detected represented as zero). D) The oxygen consumption rates from a representative experiment with control BMDMs (Ctl) and BMDMs activated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. Where not visible, error bars are obscured by the symbol. O, oligomycin; F, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP); R/A, rotenone/antimycin A (n = 1 biological with 5 technical replicates). E) ATP-Linked and maximal respiration rates for control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + P for 4 hr. (n = 4). F) Representative images of control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. Nuclei are stained with Hoechst, mitochondria are stained with MTG and membrane potential as determined by TMRE. G) Bulk membrane potential as measured by Tetramethylrhodamine, ethyl ester (TMRE) fluorescence per mitochondrial area detected by MitoTracker Green (MTG) for control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. Data is shown relative to control (n = 4). H) Representative images of control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 24 hr. analyzed via fluorescence lifetime imaging (FLIM). I) Mean endogenous NADH lifetime (𝛕mean) for whole cell measured in picoseconds (ps) from control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. Data is from one biological replicate with each data point representing one cell (n = 87–135). J) Intracellular abundances of itaconate and succinate from control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. (n = 4). K) State 3 respiration from permeabilized control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. Permeabilized BMDMs were offered pyruvate/malate, citrate, glutamate/malate, or succinate/rotenone as substrates (n = 4). L) Percent enrichment of denoted isotopologues from 13C6 Glucose from control (Ctl) and BMDMs treated with Pam3, Poly I:C, or Pam3 + Poly I:C for 4 hr. (n = 4). M&N) Pro-inflammatory gene expression for control (Ctl) and BMDMs treated with Pam3 (n = 3) (M) or Poly I:C in combination with mitochondrial effector compounds (100 nM piericidin, 30 nM antimycin A, 10 nM oligomycin, 10 nM oligomycin + 3 μM Bam15, 30 μM CAT) for 4 hr. relative to control (Ctl) (n = 3) (N). All data are mean ± SEM with statistical analysis conducted on data from biological replicates, each of which included multiple technical replicates, unless otherwise indicated. Statistical analysis for (A) and (C-D) was performed as a paired, two-tailed t-test. Statistical analysis for (F), (H), and (J-N) was performed as an ordinary one-way, ANOVA followed by Tukey’s post hoc multiple comparisons test.