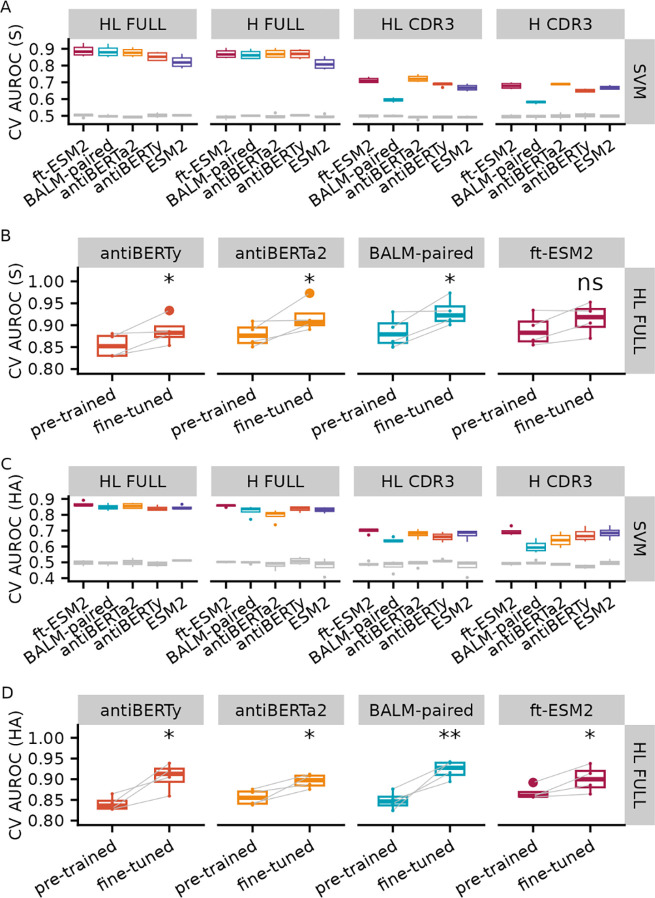
Figure 2. Specificity prediction performance for pre-trained model embedding SVM and fine-tuned antibody language models.
Box plot of 4-fold cross validation AUROC for pre-trained model embedding SVM in predicting binding to (A) SARS-CoV-2 S protein and (C) influenza HA with different sequence inputs. The gray box plots represent the random baseline by training the same model on shuffled labels. Comparison of CV AUROC between the pre-trained embedding-based SVM model and fine-tuned language models on (B) SARS-CoV-2 S protein and (D) influenza HA binding data. Each line represents the test performance for one of the CV folds. Note that the pre-trained embedding based SVM model and fine-tuned models were trained and tested on the same data for each fold. Paired t-test was used to obtain the significance level of the increase in AUROC after fine-tuning (ns: p > 0.05, *: p <= 0.05, **: p <= 0.01).